Table 3. Long tandem duplications with at least 96% identity that occur within a gene locus.
| Gene | Length | Copies | Gap | Coding |
| NBPF20 | 76181 | 2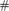
|
55 | Y |
| NBPF8 | 65137 | 2 | 0 | Y |
| CR1 | 54708 | 3 | 0 | Y |
| ANKRD30A | 47663 | 2 | 2 | Y |
| RBMY1A1 | 47081 | 3 | 0 | Y |
| NBPF12 | 44119 | 2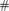
|
31 | Y |
| PGA4 | 37662 | 2 | 0 | Y |
| TRPM3 | 35986 | 2 | 0 | N |
| FCGBP | 31945 | 2
|
0 | Y |
| NEB | 31782 | 3 | 0 | N |
NKG2-E

|
30864 | 2 | 0 | Y |
| TBC1D3C/TBC1D3H | 27063 | 2 | 0 | N |
| HCAR1 | 26136 | 2 | 14 | Y |
| TTC34 | 22675 | 2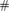
|
22 | N |
| DAZ1 | 21690 | 2 | 0 | Y |
| NBPF1 | 12620 | 2 | 17 | Y |
| NBPF12 | 12568 | 2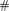
|
4 | Y |
| BRF1 | 11321 | 2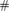
|
0 | N |
| C2orf78 | 10103 | 2 | 58 | Y |
| CLEC17A | 8924 | 2 | 0 | Y |
| TTN | 8521 | 2 | 0 | Y |
| SNTG2 | 8383 | 2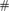
|
1 | N |
| IFI16 | 8282 | 2 | 0 | Y |
| MUC5B | 7627 | 2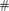
|
1 | Y |
| SPDYE3 | 7020 | 5 | 0 | Y |
| ERC1 | 5850 | 2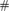
|
23 | N |
| HRNR | 5637 | 2
|
0 | Y |
| ACRC | 4289 | 2 | 66 | Y |
| SPRN | 4144 | 2 | 0 | N |
| TMEM132D | 3907 | 2
|
12 | N |
| HP/HPR | 3431 | 2 | 4 | Y |
Duplications were identified as described in the Methods. The length indicates the total length of the high-identity tandem duplicons. The gap is the separation between the two highest-identity (long) duplicons, which was required to be less than 100 bp. The duplication is “coding” if a duplicon overlaps at least one exon.
 FCGBP has a third duplicon, but with less than 96% identity.
FCGBP has a third duplicon, but with less than 96% identity.
 These genes have duplicons that are themselves STRs of lower fidelity; only the copy number for the high-identity long tandem duplication is reported in this table.
These genes have duplicons that are themselves STRs of lower fidelity; only the copy number for the high-identity long tandem duplication is reported in this table.
 The segmental duplication containing NKG2-E overlaps the three genes KLRC1, KLRC2 and KLRC3.
The segmental duplication containing NKG2-E overlaps the three genes KLRC1, KLRC2 and KLRC3.
