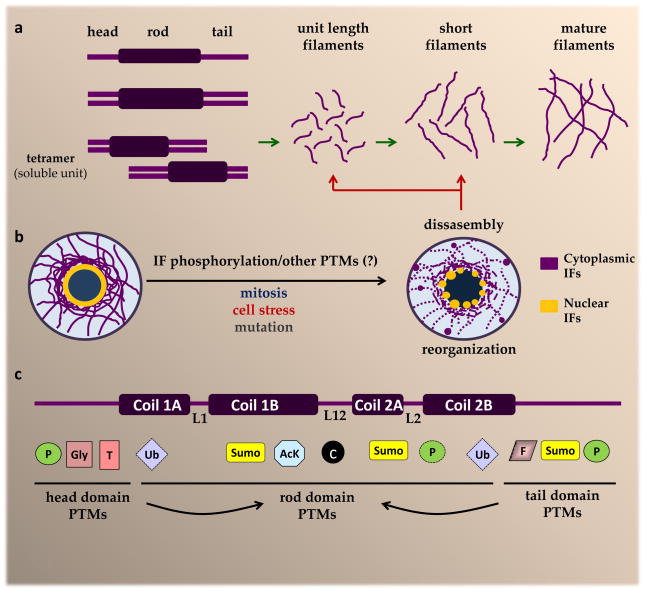
Figure 1. Cytoplasmic IF assembly and re-organization in response to phosphorylation and other PTMs.
a. The IF protein structure includes three distinct domains (head, rod, and tail)24. Cytoplasmic IFs undergo homo- or heterodimerization. The type III IFs vimentin and desmin undergo homodimerization, which is followed by anti-parallel association between homodimers to form soluble tetramers. The further assembly of vimentin and desmin filaments involves lateral association between 8 tetramers to form a unit length filament, followed by the longitudinal annealing of these into short filaments, and radial compaction of the annealed segments to yield mature IFs4. b. IF structures are highly dynamic and re-organize in certain physiologic situations, such as during mitosis, and under conditions of cell stress and in response to mutation. Site-specific phosphorylation of head and tail domain residues is a major facilitator of IF re-organization13–16, although other post-translational modifications (PTMs) are also likely to play a role. c. In addition to phosphorylation, IF proteins undergo several other modifications that target residues in the head, tail and rod domain, including the linker regions (L1, L12, L2). Rod domain PTMs are likely facilitated by head and tail domain PTMs, such as phosphorylation, as indicated by the arrows. The development of PTM-specific IF-transgenic mouse models, in combination with significant advances in proteomic technologies and antibodies targeted to specific modifications, have accelerated the pace of discovery for novel IF modifications, many of which are altered in the context of disease-causing IF mutants. AcK, lysine acetylation; c, caspase cleavage; F, farnesylation; Gly, O-linked glycosylation; P=phosphorylation; SUMO, sumoylation; T, transamidation; Ub, ubiquitination.