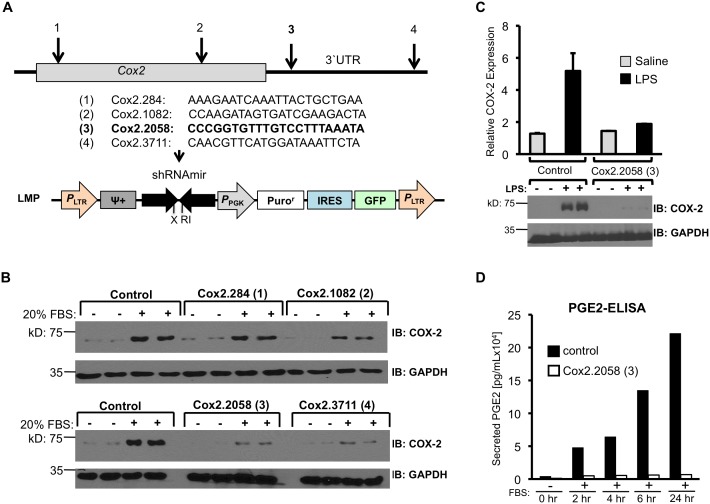
Figure 1. Identifying small inhibitory RNAs for COX-2.
(A) Schematic representation of the Cox2 transcript, indicating target areas of four designed shRNAs. The COX-2 protein-coding region is indicated as a filled box, 5′ and 3′ untranslated mRNA as solid lines. Four 22-nucleotide shRNAs for Cox2 were designed; (1) Cox2.284, (2) Cox2.1082, (3) Cox2.2058 and (4) Cox2.3711, the number reflecting the first nucleotide position of the target sequence in the mRNA transcript. The shRNA sequences were then converted into cloning templates and ligated into the LMP retrovirus vector. This vector contains an XhoI/EcoR1 cloning site for shRNAs within a miR30 backbone (shRNAmir). The LMP construct is shown as it appears after integration; shRNAs are constitutively expressed from the 5′LTR promoter. The LMP retrovirus also encodes a puromycin-resistance gene for selection and GFP as a fluorescent marker. (B) NIH3T3 cells were transduced at a low MOI with each of the four LMP vectors containing miR30-based shRNAs that target the Cox2 transcript, or with a control luciferase-targeted shRNA. Retrovirus-transduced cells were selected for integrated provirus by culturing in puromycin. Puromycin-selected cell populations were shifted overnight to 1% serum, then treated with 20% serum for 6 hours. Cell extracts were immunoblotted for COX-2 and GAPDH. (C) RAW264.7 cells stably transduced either with the luciferase shRNA LMP vector or with the LMP vector encoding Cox2-targeted Cox2.2058 shRNA were stimulated with LPS (50 ng/mL) or with saline for four hours. Cell extracts were prepared and analyzed for COX-2 protein and GAPDH. The quantified COX-2 signal was normalized against GAPDH. (D) PGE2 accumulation in the media of serum-stimulated NIH3T3 cells expressing shRNA Cox2.2058 or the control luciferase shRNA. NIH3T3 cells expressing the two shRNAs were shifted from media containing 1% FBS to 20% FBS and, at times shown, media samples were assayed for PGE2 levels.