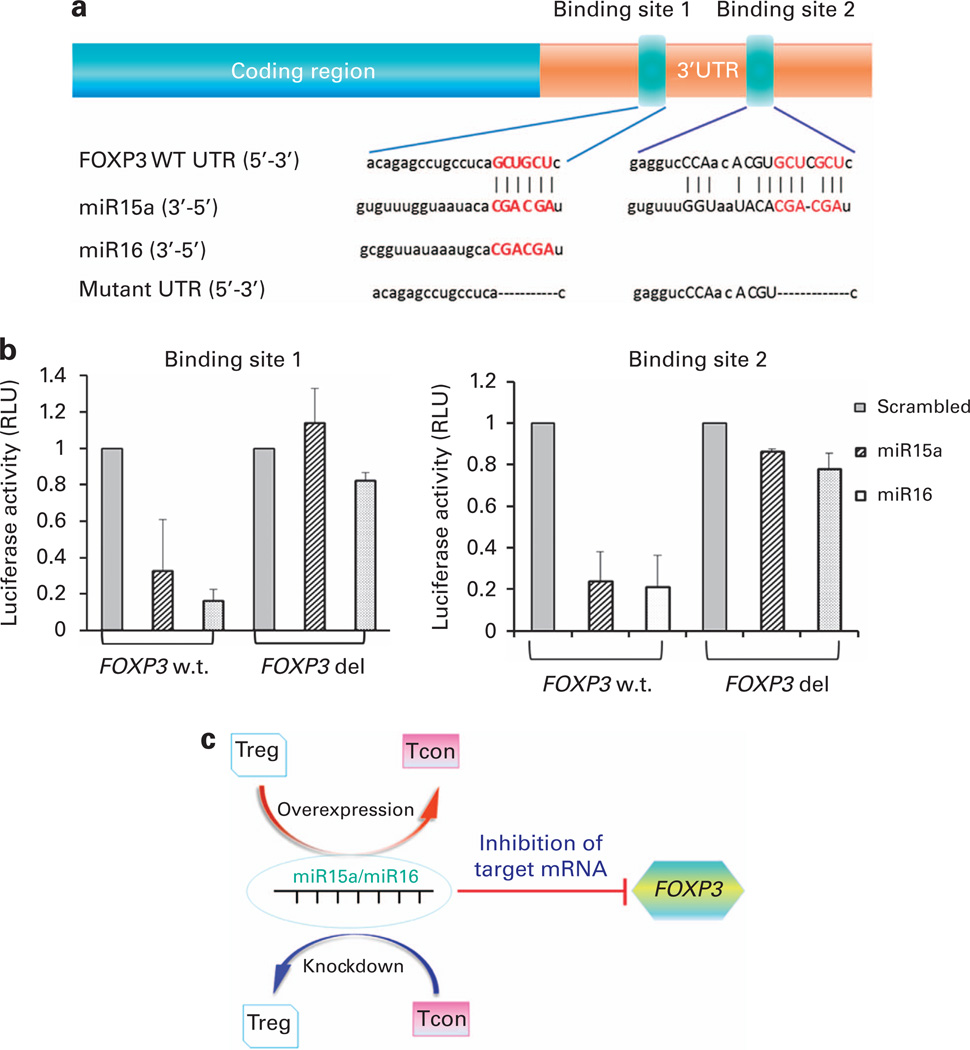
Figure 3. FOXP3 is a direct target of miR15a/16.
(a) Sequences and introduced mutations of the miR15a and miR16 binding sites. The putative target sites for the miR15a seed sequence were identified by using TargetScan25 and microRNA.org26 software. The interaction between miR15a/16 and FOXP3 was predicted by three algorithms: miranda, PITA and RNA22. Annotation of the FOXP3 3′-UTR showing predicted binding sites of miR15a and miR16, their seed sequences and subsequent introduced mutations. Two sites were identified: binding site 1: 5′-GTGGTTCTAGACACCCCCTCCCCCATCATA-3′ (forward) and 5′-GTGGTTCTAGAGGCTCTCTGTGTTTTGGGGT-3′ (reverse) and; binding site 2: 5′-GTGGTTCTAGACCTACACAGAAGCAGCGTCA-3′ (forward) and 5′-GTGGTTCTAGAGATCAGGGCTCAGGGAATGG-3′ (reverse). Using a Quik-Change site-directed mutagenesis was identified in the following primers: binding site 1, 5′-CACAGAGCCTGCCTCACGCACAGATTACTTC-3′ (forward) and 5′-GAAGTAATCTGTGCGTGAGGCAGGCTCTGTG-3′ (reverse); and binding site 2, 5′-GAGGTCCCAACACGTCACACACACGGCCTG-3′ (forward) and 5′-CAGGCCGTGTGTGTGACGTGTTGGGACCTC-3′ (reverse). (b) miR15a/16 directly interact with Foxp3 at binding site 1 and binding site 2. In luciferase-based assay, miR15a/16 showed direct binding interaction with FOXP3 at binding site 1 and binding site 2. When these sites are specifically mutated, mir15a/16 lose their ability to bind to FOXP3. (c) ‘Toggle-switch’ role of miR15a/16 and FOXP3 function. Overexpression of miR15a/16 in Tregs can push their functional differentiation into a more conventional T-cell-like activity. On the other hand, silencing of miR15a/16 pathway in Tcon may lead to an inducible Treg-like function.