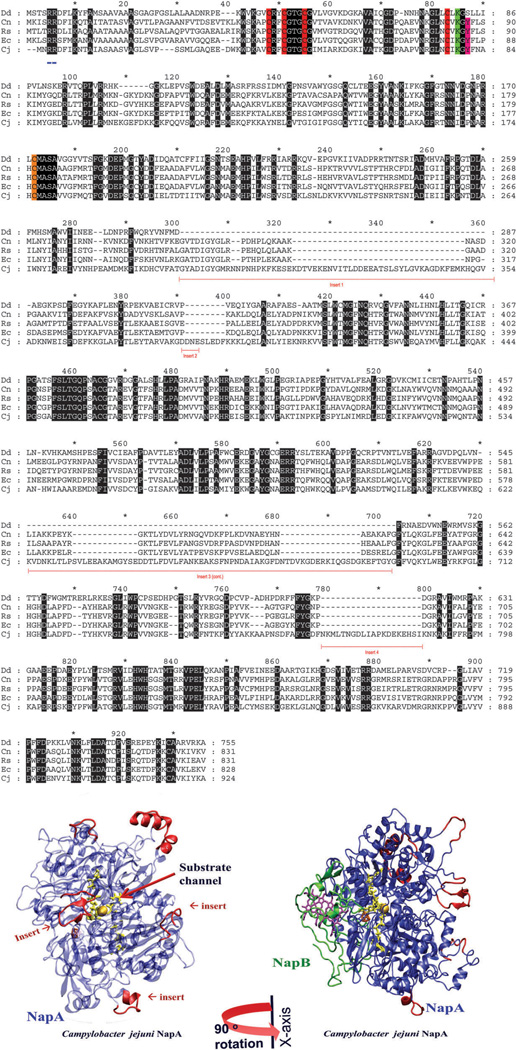
Fig. 8.
A sequence alignment, created using GeneDoc, of NapA from D. desulfuricans, Ralstonia eutropha, Rhodobacter sphaeroides, E. coli, and C. jejuni, representing δ, α, β, γ and ε proteobacteria, respectively. Sequence inserts (underlined in red), identical residues (black), [4Fe–4S] binding motif (red), the Mo-coordinated cysteine residue (orange); the lysine (green) and tyrosine (pink) are shown here, the latter two residues are predicted to be involved in electron transfer. The twin arginine translocase (TAT) signal peptide is underlined in blue. Also shown (bottom panel) is the homology models of C. jejuni NapA and NapAB. The insertion sequences, shown in red, are opposite to the NapB binding site. The homology models were created using Molecular Operating Environment (MOE) software heme is purple, [4Fe–4S] is orange, and the MPT is yellow.