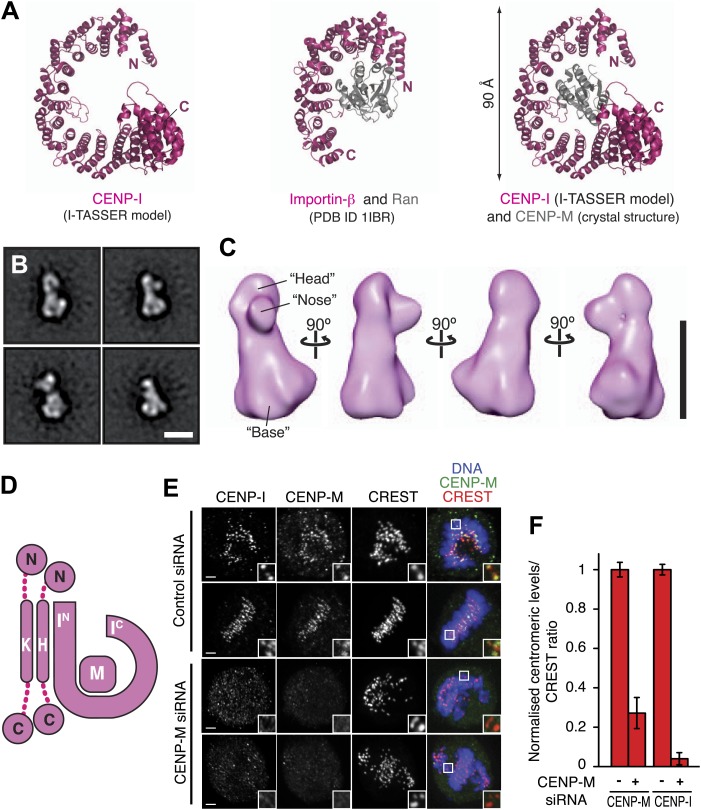
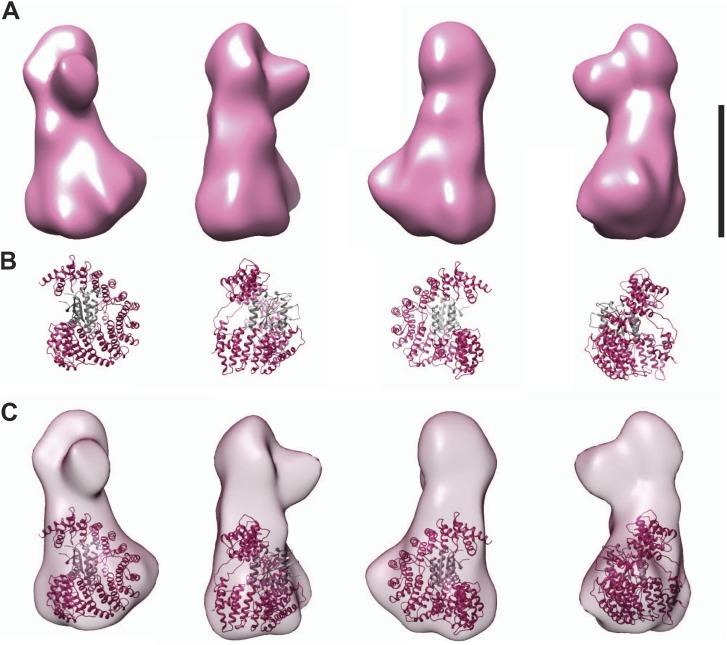
Figure 4. Structural organization of the HIKM complex.
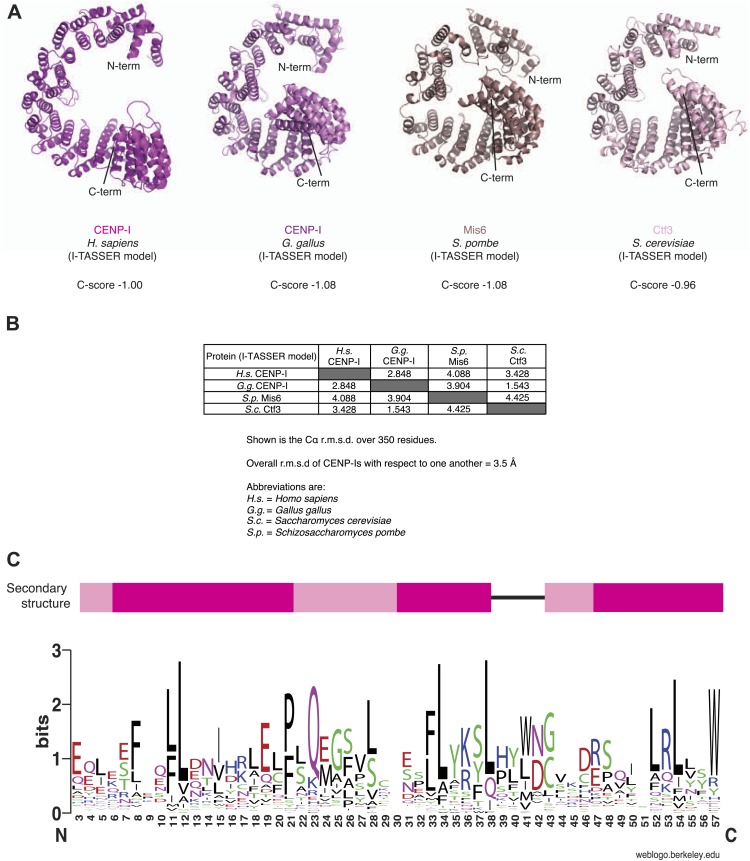
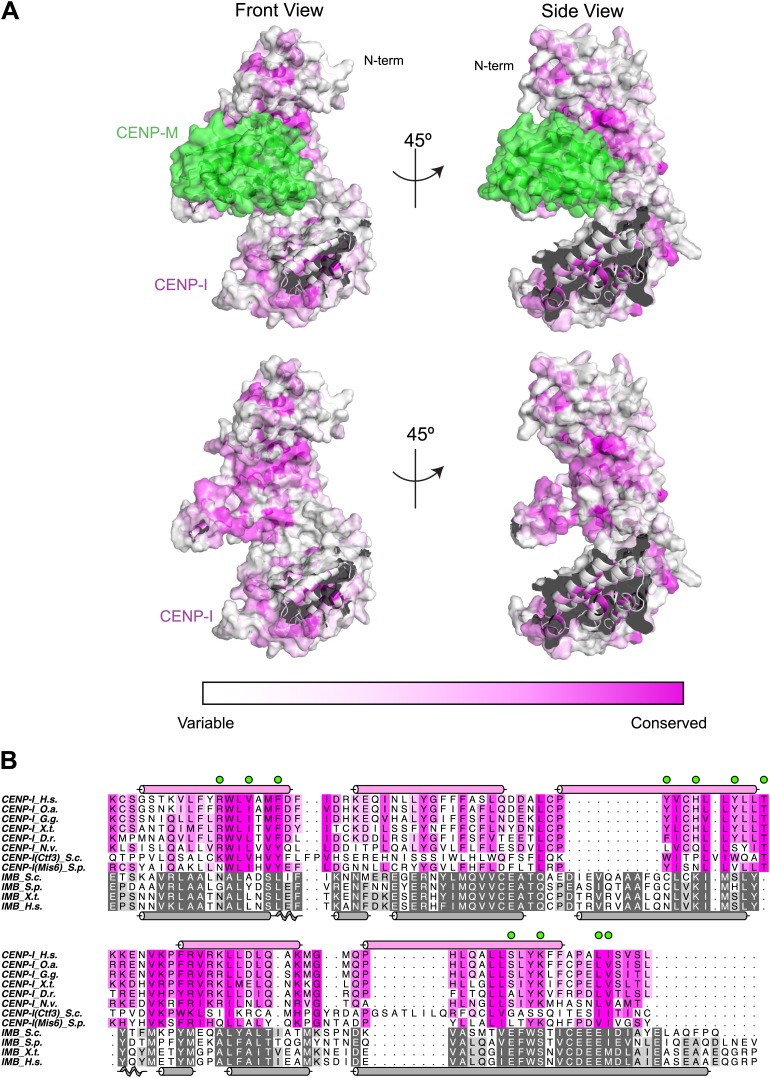
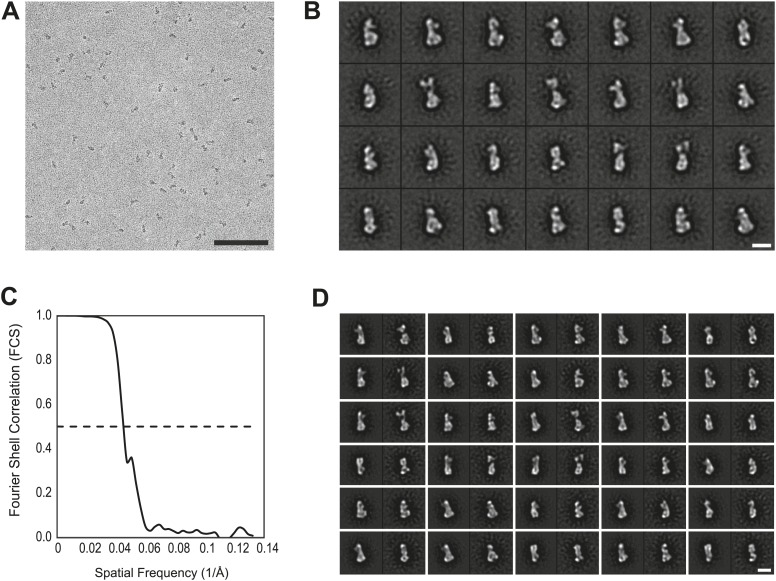
(A) Cartoon representation of the CENP-I model generated by program I-TASSER (left), of the Importin-β/Ran complex (middle), and of a hypothetical structure between CENP-I and CENP-M modeled on the Importin-β/Ran complex (right). A scoring function (C-score) associated with I-TASSER models estimates accuracy of structure predictions. C-score is typically in a range from −5 to 2, where a higher score reflects a model of better quality. Both false positive and false negative rates are estimated to be below 0.1 when a C-score >−1.5 is displayed (Zhang, 2008). The CENP-I model is associated with a C-score of −1. (B) Representative class averages of the negatively stained HIKM complex. Figure 4—figure supplement 3 shows the complete set of class averages. Scale bar = 10 nm. (C) A 3D reconstruction of HIKM complex from negatively stained particles at ∼22 Å resolution. Scale bar = 10 nm. (D) Summary of interactions in the CENP-HIKM complex. The central regions of CENP-H and CENP-K may form an extended parallel interaction, possibly through an α-helical arrangement, which interacts more or less co-linearly with the N-terminal region of CENP-I (IN). Additional globular domains may be present at the N- and C-termini of CENP-H and CENP-K. The entire sequence of CENP-I may fold as a helical solenoid. CENP-M does not interact with CENP-H/K and may bind near the concave surface of the predicted CENP-I solenoid, becoming largely buried. (E) siRNA depletion of endogenous CENP-M abrogates CENP-I kinetochore localization in HeLa cells. Representative cells displayed here are the same shown in Figure 1D, but with addition of CENP-I staining (left panels). Insets display a higher magnification of regions outlined by white boxes. Scale bars = 2 µm. (F) CENP-M and CENP-I kinetochore levels from the experiment illustrated in E. Quantification for CENP-M kinetochore levels are the same shown in Figure 1D and were performed as previously described. Graphs and bars indicate mean ± SEM.