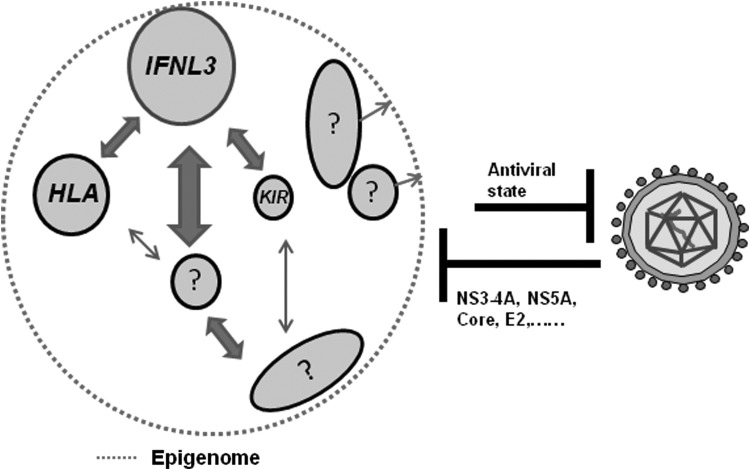
FIG. 5.
Host-virus-environment interactions determine the outcome of HCV infections. On the left is shown the immunome that is a subset of the larger host genome and on the right is shown HCV with its genome enclosed in a protein cover. The components of the immunome are shown as genes (circles) or pathways (ellipses). The size of the circle or ellipse is representative of the relative importance in the immunome. The arrows in between genes and pathways refer to genetic and physiological interactions between the components. The size and length of the arrows represent the relative importance and physical location on the genome respectively. The identified genes relevant to clearance of HCV infection are shown (such as IFNL3, HLA, and KIR, Smith and others 2011) and other yet unidentified or not validated ones are shown with question marks. The epigenome shown as a dotted line denotes the pervading influence of epigenetic (mostly transcriptional) regulation by means of alterations in the chromatin state throughout the immunome. The genes and pathways that influence (arrows pointing toward dotted line) the epigenome relevant to “HCV immunome” are not known, hence shown with question marks. The combined effect of the immunome (including innate and adaptive components) will lead to an antiviral state that will inhibit HCV replication and eliminate it from the host. On the other hand, HCV can influence the host immunome, including the epigenome. The viral factors that are known to affect the immunome (such as NS3-4A cleaving MAVS and TRIF and inhibiting innate immune signaling, Lemon 2010; core affecting DNA methylation, Lee and others 2013) are shown.