Figure 3.
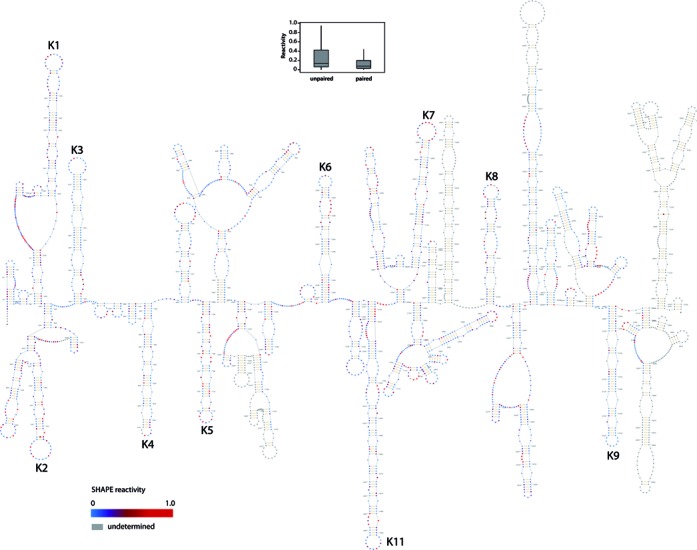
Secondary structure of KSHV pri-miR-K10/12. The secondary structure of pri-miR-K10/12, containing the intronic clustered pre-miRNAs (from pre-miR-K1 to -K9 and pre-miR-K11, as indicated in black), was determined in solution by SHAPE method. The 3157 nt-long RNA we used for the analysis was in vitro transcribed. Residues are colored with a gradient of blue to red according to the intensity of SHAPE reactivity (low to high, respectively). Nucleotides in gray were not determined. Box plots represent the SHAPE reactivity in function of the unpaired or paired fate of nucleotides in the 2D structure model. Paired nucleotides show a less scattered pattern than the one for unpaired nucleotides. Moreover median is smaller for paired than unpaired nucleotide.
