Figure 4.
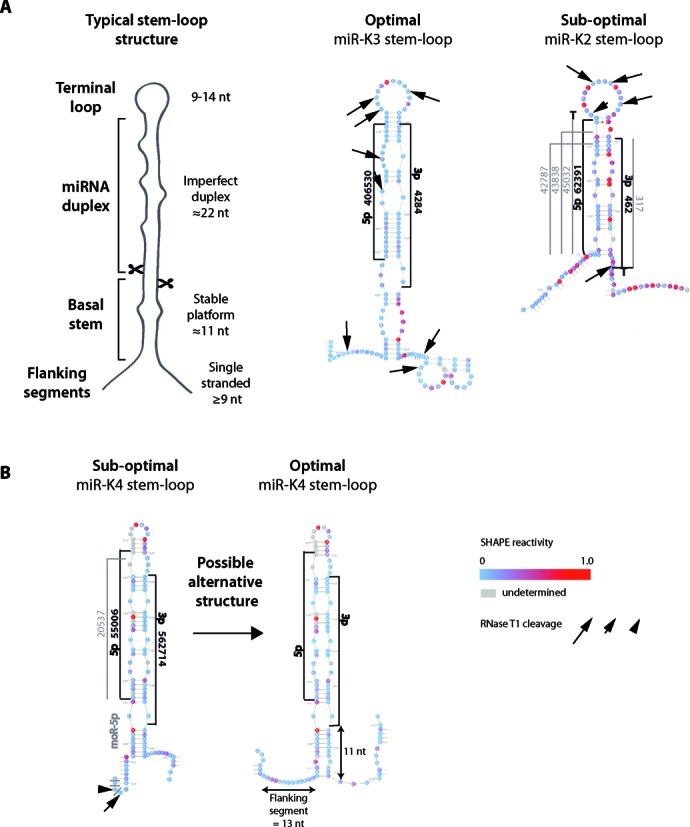
Secondary structure of typical and representative KSHV miRNA stem-loops. (A) On the left is depicted a typical miRNA stem-loop structure, with the different modules emphasized (see main text for details). miR-K3 and miR-K2 stem-loops (respectively optimally and suboptimally structured), composed of the pre-miRNA plus 20 nucleotides before and after the Drosha cleavage site, are represented as examples of optimal and suboptimal hairpin structures. SHAPE reactivity of each nucleotide is indicated. Arrows whose sizes are proportional to enzymatic activity indicate RNase T1 cleavages. The most abundant sequences of 5p and 3p arms of the miRNAs and the different isoforms are indicated in black and in gray, respectively, along with the number of sequence reads obtained by deep sequencing. Non-templated nucleotides found at the ends of the miRNA are shown. (B) In the case of miR-K4 stem-loop, manual folding allowed for an optimal alternative structure.
