Figure 1.
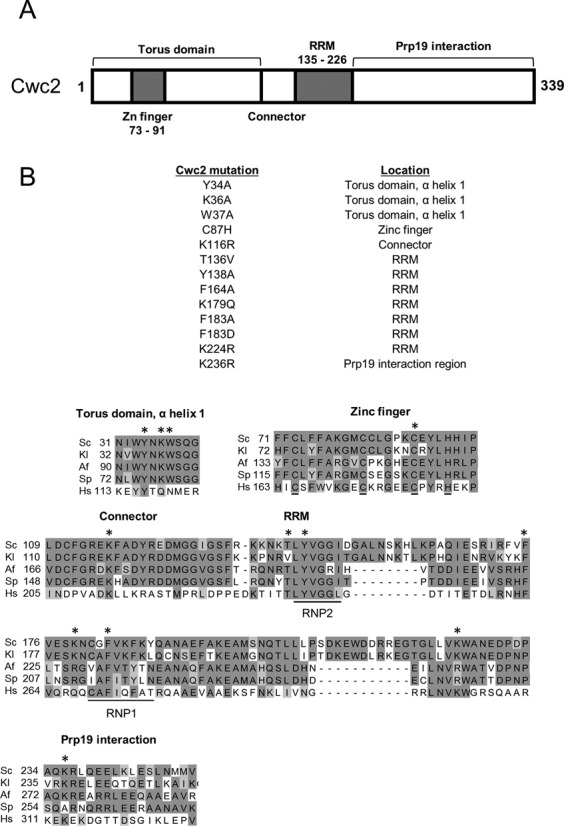
Domain structure and viable mutations in Cwc2. (A) The location of known protein motifs in the Saccharomyces cerevisiae Cwc2 protein are depicted including the zinc (Zn) finger, Torus domain, connector, RNA recognition motif (RRM) and Prp19 interaction domain. (B) Viable mutations in Cwc2 used for genetic interaction analysis. The locations of each mutation in the Cwc2 protein structure are described to the right of each mutation. The amino acid sequence of Cwc2 from Saccharomyces cerevisiae (Sc, NCBI-GI: 6319992) was aligned with orthologs from Kluyveromyces lactis (Kl, NCBI-GI: 50305857), Aspergillus fumigatus (Af, NCBI-GI: 70998470), Schizosaccharomyces pombe (Sp, NCBI-GI: 19114249) and Homo sapiens (Hs, NCBI-GI: 8922328). The locations of the Cwc2 mutants are identified with asterisks above the selected regions of the alignment.
