Figure 1.
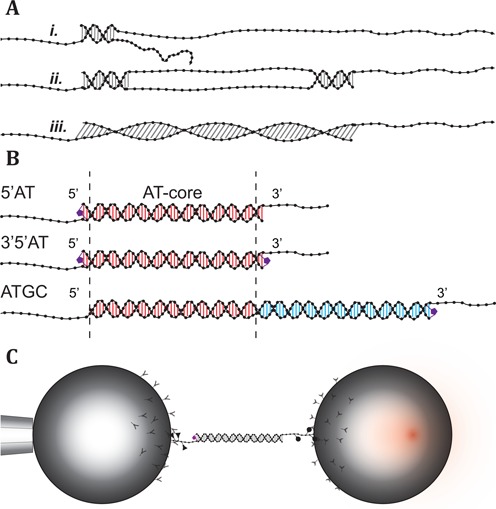
Overstretching mechanisms, DNA constructs and experimental setup. (A) The three proposed mechanisms for overstretching. (i.) Peeling, where the DNA progressively melts from free ends. (ii.) Internal melting, where the melting is initiated in regions of low stability forming ‘bubbles’. (iii.) B-to-S transition, where the base-pairing remains intact as the duplex is extended into a longer form. (B) Schematic description of the DNA constructs and the covalent inter-strand linkage. 5′AT: 64 bp with a terminal inter-strand cross-link. The inter-strand linker is formed by joining alkyne and azide modified bases on opposite strands together using the Cu+-catalyzed click reaction. 3′5′AT: 64 bp with one inter-strand linker at each end of the duplex region. ATGC: 122 bp duplex region consisting of the AT-core sequence shared with 5′AT and 3′5′AT, and a previously studied single-clicked GC-rich sequence (23). For bead attachment, each construct is extended in the 3′-ends with single-stranded DNA containing digoxigenin or biotin modified bases. (C) Experimental setup. The DNA constructs are tethered to the streptavidin and anti-digoxigenin coated beads by biotin and digoxigenin modified bases incorporated in the single stranded handles, respectively. One of the beads (left) is immobilized by suction onto a pipette, while the other (right) is manipulated by the optical trap.
