Figure 12.
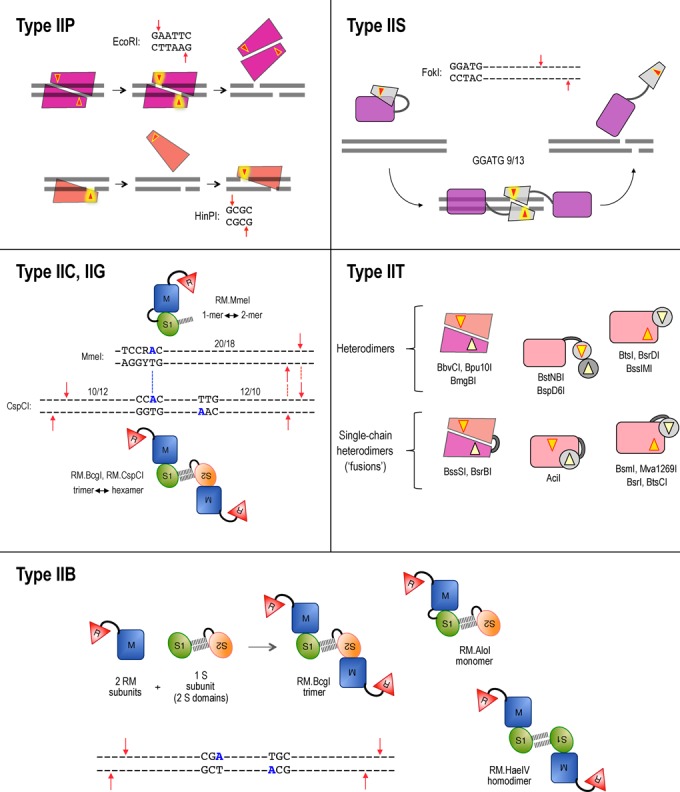
The subunit composition and cleavage mechanism of selected subtypes of Type II REases. Type IIP enzymes act mainly as homodimers (top), and cleave both DNA strands at once. Some act as dimers of dimers (homotetramers) instead, and do the same. Still others act as monomers (bottom) and cleave the DNA strands separately, one after the other. Bright triangles represent catalytic sites. Type IIS enzymes generally bind as monomers, but cleave as ‘transient’ homodimers. Type IIB enzymes cleave on both sides of their bipartite recognition sequences. Their subunit/domain stoichiometry and polypeptide chain continuity varies. Three examples of primary forms are shown: BcgI, AloI and HaeIV. These forms assemble in higher-order oligomers for cleavage. Type IIB enzymes display bilateral symmetry with respect to their methylation and cleavage positions. It is not clear whether they cleave to the left or to the right of the half-sequence bound. Type IIG enzymes (e.g. BcgI) might cleave upstream (left) of their bound recognition half-site. All other Type IIG enzymes (e.g. MmeI) cleave downstream from the site, often with the same geometry. These proteins have very similar amino acid sequences, however, suggesting that somehow the reactions are the same. Type IIT enzymes cleave within or close to asymmetric sequences. Composition varies; they have two different catalytic sites: top-strand specific and bottom-strand specific. In some, both subunits/domains interact with the recognition sequence (left cartoons). In others, only the larger subunit/domain recognizes the DNA.
