Figure 6.
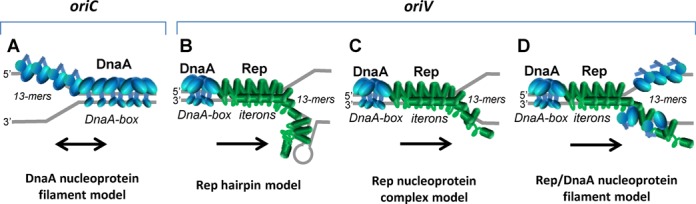
Alternative models of nucleoprotein complex formation at the origin region by bacterial and plasmid replication initiators. In the presented models the regions of oriC and oriV origins, and initiator proteins, DnaA (blue) and TrfA (green) are marked. (A) Model of binding of the DnaA protein (blue) to dsDNA containing DnaA-boxes and to ssDNA DUE containing three 13-mers. DnaA-ATP forms a filament at the DnaA-boxes. Such filament extends to the 13-mers at the ssDNA. The model, based on Duderstadt et al. (31), presents only the AAA+ and DBD domains of DnaA. (B–C) Alternative models of binding of the plasmid RK2 replication initiator TrfA protein at the dsDNA containing iterons and the ssDNA DUE containing the four 13-mers. (B) TrfA protein binding to the iterons results in DUE melting. The melted ssDNA forms local secondary structures that are further stabilized by the TrfA. This process is accompanied by DnaA protein bound to four DnaA-boxes. (C) TrfA protein binds to iterons which results in DUE melting. Then, it extends along one of the two single strands of DNA. The DnaA protein bound to four DnaA-boxes is shown in blue. (D) The coordinated binding of TrfA protein to iterons and the DnaA protein to DnaA-boxes causes melting of the DUE. Then the TrfA and DnaA could bind to the same DNA strand forming a heterofilament or each of the proteins could bind to different strands. The orientation of the origin models is as presented by Duderstadt et al. (31) (A) and Kowalczyk et al. (8) (B–D). Black arrows in the illustrations indicate the direction of replication. The proposed models do not exclude discontinuity of Rep protein filaments, neither aim to state the nature or structure of tripartite complexes (see the discussion for details).
