Figure 5.
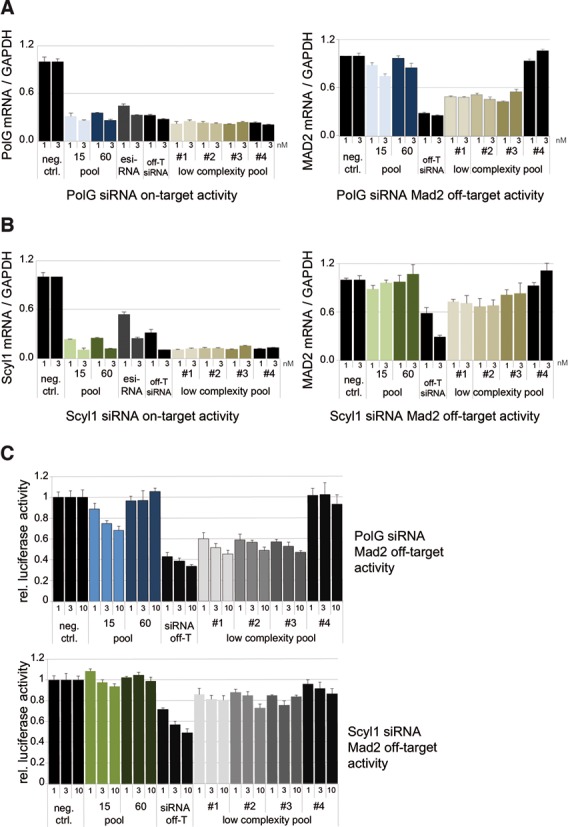
Comparison of siPools with other available RNAi reagents. qPCR analysis of on- (left panels in (A) and (B)) and off-target (right panels in (A) and (B)) activities of various siRNA tools. HeLa cells were transfected with 1 or 3 nM siPool 15, siPool 60, single off-T siRNA, esiRNAs and four different low-complexity pools directed against PolG (A) or Scyl1 (B). mRNA levels were normalized to GAPDH and relative expression levels were calculated using a negative control siRNA. Low-complexity pool #4 served as a MAD2 off-target negative control. (C) HeLa cells were transfected with 1, 3 or 10 nM siRNA off-T, siPools with 15 or 60 different siRNAs and four different low-complexity pools directed against PolG (blue) or Scyl1 (green). Off-target activity was analyzed using a reporter system based on firefly-luciferase activity controlled by the MAD2 3′-UTR. Relative luciferase activity was calculated using the ratio of firefly/Renilla luciferase and via normalization to the corresponding ratios of the empty control vector. Low-complexity pool #4 served as a MAD2 off-target negative control.
