Figure 2.
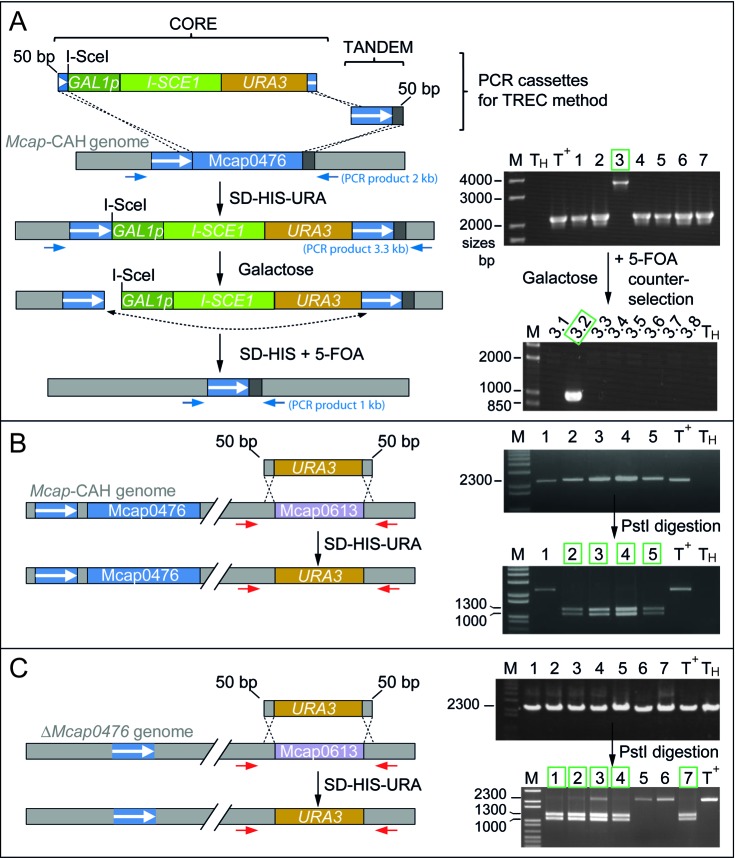
Inactivation of the Mcap0476 and/or Mcap0613 genes in Mcap genomes maintained in yeast. (A) Strategy for inactivation of Mcap0476 using the TREC method (30) whereby two overlapping cassettes (CORE and TANDEM) containing Mcap0476 sequences were introduced into yeast spheroplasts carrying the Mcap-CAH genome. Yeast recombinants were selected on solid SD-HIS-URA medium. Homologous recombination with three crossing-over events formed direct tandem repeats (white arrow, blue background) and resulted in the replacement of Mcap0476 by the CORE and TANDEM cassettes. Replica-plating of yeast recombinants on galactose solid medium induced expression of I-SceI followed by cleavage at the I-SceI site. This double-strand break increased the frequency of homologous recombination events (dash line) between the tandem repeats followed by cassette excision, which was selected for by plating on solid medium containing 5-fluoroorotic acid (SD−HIS + 5-FOA). Yeast recombinants were analyzed using the PCR primer pair TrmFO15a and TrmFO16 (Supplementary Table S1) positioned on either side of Mcap0476 (blue arrows). PCR products of 2 kb showed that Mcap0476 was still intact in clones 1, 2, 4, 5, 6 and 7, while the 3.3 kb band from clone 3 revealed the desired insertion of CORE and TANDEM cassettes. Subsequent excision of the cassettes is indicated by the 1 kb PCR band from subclone 3.2. This subclone, renamed W303-ΔMcap0476, was analyzed further by multiplex PCR and PFGE (Supplementary Figure S3). (B) Mcap0613 was deleted in Mcap-CAH by conventional gene replacement with a URA3 cassette. Yeast spheroplasts housing the Mcap-CAH genome were transformed with the cassette to promote homologous recombination involving two cross-overs and resulting in the replacement of Mcap0613 by URA3. PCR reactions were performed on candidate yeast clones with the primer pair TrmFO25b and TrmFO26 (red arrows) (Supplementary Table S1). The URA3 and Mcap0613 genes both produce fragments of 2.3 kb, and were discriminated by cleavage at PstI in URA3 into 1.0 and 1.3 kb bands. Positive W303-ΔMcap0613::URA3 clones (2, 3, 4 and 5) are boxed in green. (C) The same procedure was applied to the W303-ΔMcap0476 strain to produce the double knockout W303-ΔMcap0476/ΔMcap0613::URA3. Positive clones (1–4 and 7) are boxed in green. Positive clones were taken further for multiplex PCR and PFGE analyses (Supplementary Figure S3). M, DNA size ladder (Invitrogen); T+, DNA from Mcap-wt cells; TH, negative control, no DNA.