Figure 4.
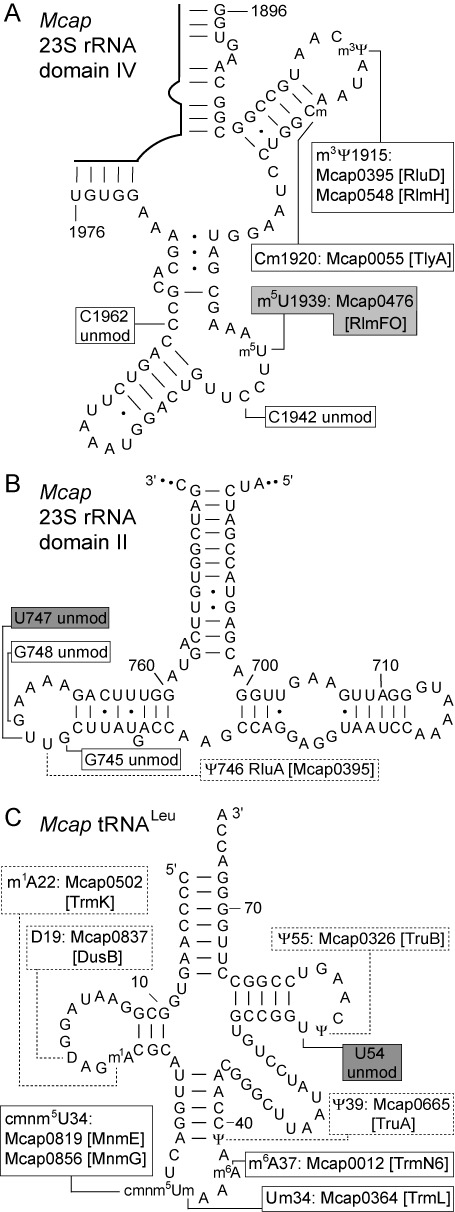
Mcap RNA secondary structures showing the modification sites. The positions of m5U modification known from other organisms are indicated (gray boxes). (A) Region of Mcap 23S rRNA domain IV with the RlmFO (Mcap0476) methyltransferase product m5U1939. Modifications at other nucleotides were evident during this study, and are indicated (white boxes) together with the putative Mcap enzymes (15). Nucleotides 1942 and 1962 are unmodified in Mcap rRNA (unmod) whereas m5C is found at these positions in some bacteria (49,50). (B) In certain bacteria, methylation occurs at the conserved nucleotides G745, U747 and/or G748 (3,51,52). There was no methylation in this region of Mcap 23S rRNA domain II (Supplementary Figure S8) and, consistently, Mcap possesses none of the orthologous methyltransferases. (C) Structure of the Mcap tRNALeu UAA isoacceptor with nucleotide modifications (24). We confirmed the absence of modification at U54 using HPLC (Supplementary Figure S6), and the anticodon loop modifications were consistent with our MS (Supplementary Figure S7) and bioinformatics analyses. Modifications in the dashed boxes were not tested empirically in the present study, but were supported by bioinformatics.
