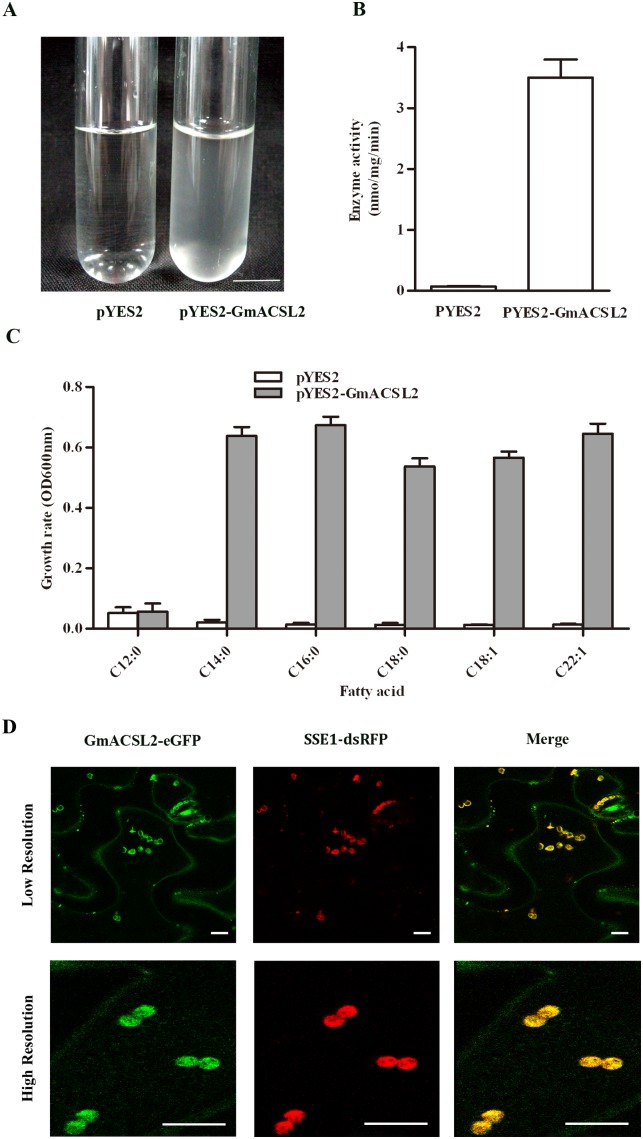
Figure 2. Yeast complementation test and subcellular localization.
(A) Yeast complementation test. Right: culture of yeast strain YB525 cells containing the pYES2 empty plasmids; Left: culture of yeast strain YB525 cells containing the pYES2-GmACSL2 plasmids. Bar = 1 cm. (B) ACS enzyme activities. YB525 cells carrying the pYES2 or pYES2-GmACSL2 plasmids were harvested after galactose induction for 18 h. 1-[14C] oleic acid was used as a substrate. Enzyme activities were measured based on the [14C] label incorporated into the acyl-CoA fraction per assay. Values are means of triplicate with standard deviation (SD). (C) Growth rate of the transformed line in different fatty acid culture medium. YB525 was transformed with the pYES2-GmACSL2 and empty PYES2 plasmids, which were cultured in liquid medium with various fatty acids (12∶0, 14∶0, 16∶0, 18∶0, 18∶1, and 22∶1) as the sole carbon source. (D) Subcellular localization. Fluorescence signals of eGFP were detected in cells expressing GmACSL2-eGFP fusion protein by Leica TCS scanning confocal microscope (left panels). Fluorescence signals of dsRFP were detected in cells expressing SSE1-dsRFP fusion protein (middle panels). Right panels were merged by left and middle panels. The upper panels are low-resolution pictures and the lower panels were high-resolution pictures. The immunofluorescence was done with tobacco leafs. Bar = 10 µm. Data are presented as the mean ± SEM of three experiments.