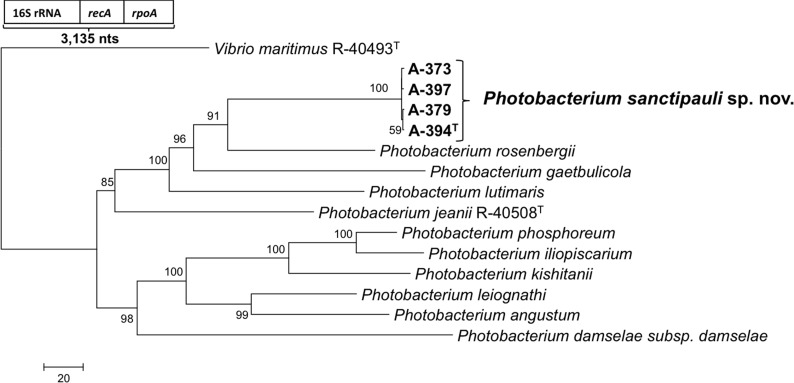
Figure 2.
Neighbour-joining phylogenetic tree based on concatenated 16S rRNA, recA and rpoA gene sequences (3,135 nt) showing the position of P. sanctipauli sp. nov. The evolutionary distances were computed using the number of differences method and are in the units of the number of base differences per sequence. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (Pairwise deletion option). Phylogenetic analyses were conducted in MEGA5. Bootstrap values (>50%) based on 1,000 resamplings are shown. Vibrio maritimus R-40493T was used as outgroup. Bar estimate sequence divergence.