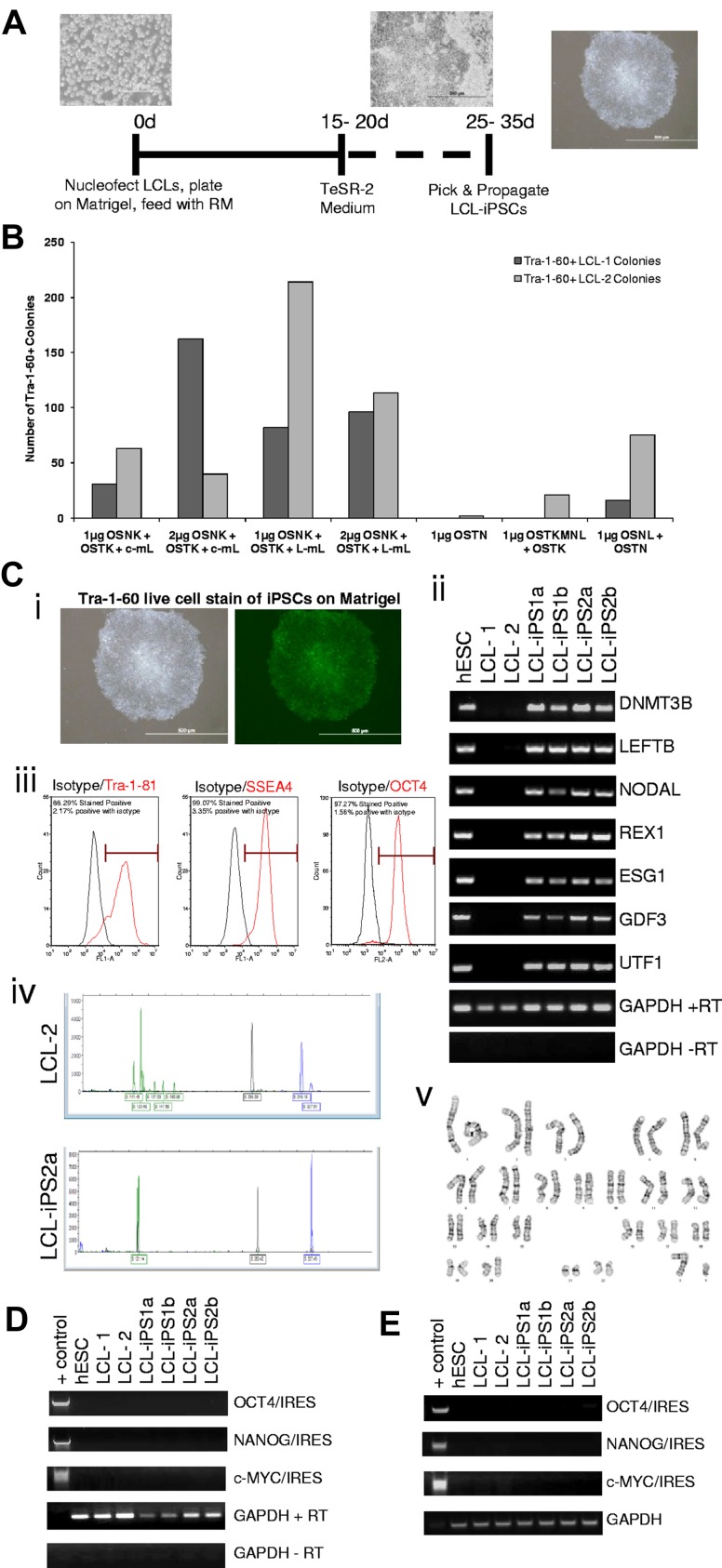
Figure 1.
Derivation and characterization of LCL-iPSCs. (A) The scheme for reprogramming human LCLs. Morphology of an LCL culture, cells during reprogramming, and LCL-iPSC colonies. Images were captured using original magnification ×40, ×20, and ×10, respectively. (B) The graph illustrates the average number of Tra-1–60-positive colonies formed after nucleofection of LCL-1 and LCL-2 with various combinations of reprogramming factors at day 40 after nucleofection. The abbreviations used for the reprogramming factors are as follows: O indicates OCT4; S, SOX2; N, NANOG; K, KLF4; c-m, c-MYC; L-m, L-MYC; T, SV40 large T antigen; and L, LIN28. Construction of the plasmids and the emergence of the colonies are described in the Supplemental data. (C) Characterization of LCL-iPSC lines. (i) Brightfield (left panel) and Tra-1–60 staining (right panel) of a representative LCL-derived iPSC line (LCL-iPS2a) confirms uniform expression of the pluripotent surface marker, Tra-1–60. Images were taken using original magnification ×10. (ii) RT-PCR analysis of H1 (hESC line), LCL-1, LCL-2, and 2 representative LCL-iPS clones from each donor lines for expression of hES cell-marker genes DNMT3B, LEFTB, NODAL, REX1, ESG1, GDF3, and UTF1. GAPDH was used as positive loading control for each sample, and cDNA made in the absence of reverse transcriptase (no RT) was used to verify that genomic DNA did not contaminate the RNA samples. (iii) Flow cytometric analysis of hESC pluripotency markers Tra-1–81 (left panel: red line indicates Tra-1–81; and black line, isotype control), SSEA-4 (middle panel: red line indicates SSEA4; and black line, isotype control), and OCT4 (right panel: red line indicates OCT4; and black line, isotype control) of a representative LCL-derived iPS line, LCL-iPS1b. (iv) Each B-cell has a respective single productive immunoglobulin gene rearrangement that is unique in both length and sequence. PCR assays with specific primers for the joining region and all 3 of the conserved framework regions (FR1, FR2, and FR3) were used to amplify immunoglobulin heavy chain (IgGH) gene rearrangements according to the BIOMED-2 Concerted Action protocol. DNA from a normal or polyclonal B-cell population produces a bell-shaped curve of amplicon products (Gaussian distribution), whereas clonal rearrangements are identified as prominent, single-sized products by capillary electrophoresis and GeneScanning. (v) Karyotype analysis of each iPSC line was performed after 11 passages and found to be normal (a representative image of LCL-iPS2b is shown). (D) RT-PCR analysis of RNA from LCL-iPSCs for the expression of reprogramming vector genes at passage 25. Forward primers for 3 reprogramming genes of interest (OCT4, NANOG, and c-MYC) and reverse primers for internal ribosome entry site 2 were used. GAPDH was used as a positive loading control for each sample, and cDNA made in the absence of reverse transcriptase (no RT) was used to verify that genomic DNA did not contaminate the RNA samples. A reprogramming vector was included as a positive control for each PCR reaction (+ control). (E) PCR analysis of genomic DNA confirms no integration of the transgenes in LCL-iPSCs at passage 25. Forward primers for 3 reprogramming genes of interest (OCT4, NANOG, and c-MYC) and reverse primers for the internal ribosome entry site were used. GAPDH was used as a positive loading control for each sample, and a reprogramming vector was included as a positive control for each PCR reaction (+ control). All images were captured on a Olympus 1×71 microscope equipped with a Olympus DP70 camera and DP Controller Version 3 software.