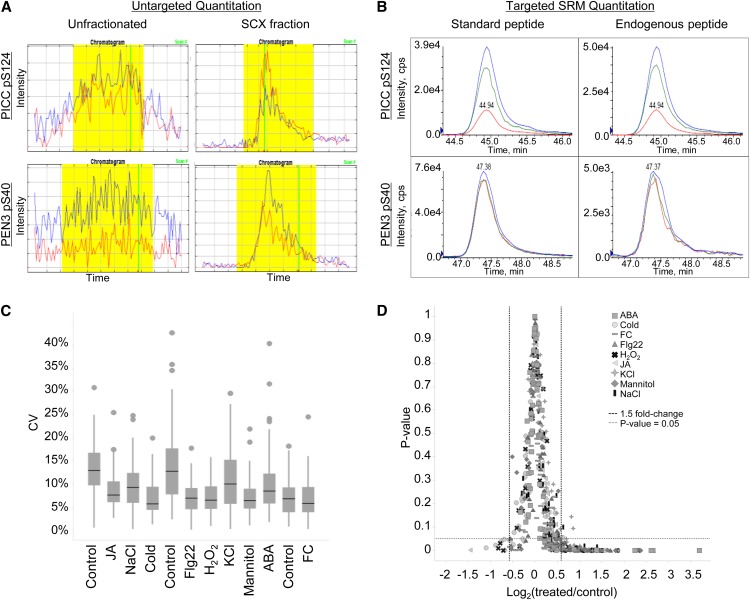
Figure 5.
Improved phosphopeptide detection using targeted SRM analysis. A and B, Quantification of the PAMP-Induced Coiled-Coil (PICC) phosphopeptide DIDLSFS[pS]PTK (top) and the PEN3 phosphopeptide NIEDIFSSG[pS]R (bottom) across different MS platforms. A, Extracted ion chromatograms from phosphopeptide enrichment of unfractionated, total protein extract (left) compared with phosphopeptide enrichment of SCX-fractionated samples (right) using untargeted proteomic methods. The PEN3 peptide was only identified in one experimental sample in SCX-fractionated data. Extracted ion chromatograms were generated by Census software. Blue lines indicate 14N peptide, and red lines indicate 15N peptide. Yellow shading indicates the area used to calculate the peptide abundance ratio in Census. Vertical green lines indicate the scan at which MS/MS data were acquired. B, Extracted ion chromatograms from phosphopeptide enrichment of unfractionated, total protein extract using the targeted SRM method. Peptide standards (left) and endogenous peptides (right) coelute with identical fragment ion patterns. Three fragment ions were measured for each peptide. C and D, Statistical analysis of targeted proteomic data. C, Box plot of the coefficient of variance (CV) for each phosphopeptide quantified across three biological replicates in 12 different treatments. A total of 58 phosphopeptides are plotted in each treatment. The control treatment is presented three times because samples were grown and processed in three separate batches. Black bars indicate the median value for each treatment. D, Phosphorylation changes of 1.5-fold or greater are statistically significant (P ≤ 0.05, Student’s t test) in almost all targeted experiments. FC, Fusicoccin; JA, methyl jasmonate.