An Arabidopsis zinc finger protein acts as a negative regulator of ABA-suppressed seed germination and modulates plant development, fertility, and hypocotyl elongation under red light.
Abstract
Seed germination is controlled by environmental signals, including light and endogenous phytohormones. Abscisic acid (ABA) inhibits, whereas gibberellin promotes, germination and early seedling development, respectively. Here, we report that ZFP3, a nuclear C2H2 zinc finger protein, acts as a negative regulator of ABA suppression of seed germination in Arabidopsis (Arabidopsis thaliana). Accordingly, regulated overexpression of ZFP3 and the closely related ZFP1, ZFP4, ZFP6, and ZFP7 zinc finger factors confers ABA insensitivity to seed germination, while the zfp3 zfp4 double mutant displays enhanced ABA susceptibility. Reduced expression of several ABA-induced genes, such as RESPONSIVE TO ABSCISIC ACID18 and transcription factor ABSCISIC ACID-INSENSITIVE4 (ABI4), in ZFP3 overexpression seedlings suggests that ZFP3 negatively regulates ABA signaling. Analysis of ZFP3 overexpression plants revealed multiple phenotypic alterations, such as semidwarf growth habit, defects in fertility, and enhanced sensitivity of hypocotyl elongation to red but not to far-red or blue light. Analysis of genetic interactions with phytochrome and abi mutants indicates that ZFP3 enhances red light signaling by photoreceptors other than phytochrome A and additively increases ABA insensitivity conferred by the abi2, abi4, and abi5 mutations. These data support the conclusion that ZFP3 and the related ZFP subfamily of zinc finger factors regulate light and ABA responses during germination and early seedling development.
The phytohormone abscisic acid (ABA) controls seed development, germination, and responses to dehydration caused by either drought or high soil salinity. ABA regulates seed maturation, promotes seed dormancy and desiccation tolerance and the synthesis of seed storage proteins and lipids, and inhibits phase transitions from embryonic growth to germination and from vegetative to reproductive growth. In addition to its role in plant development, ABA controls responses to drought and salt stresses, including stomatal closure, activation of genes involved in osmotic adjustment, ion compartmentalization, regulation of shoot versus root growth, and modifications of root hydraulic conductivity (Verslues and Zhu, 2005; Sirichandra et al., 2009; Fujita et al., 2011; Finkelstein, 2013).
In the last decades, numerous genes controlling ABA biosynthesis and signal transduction were identified by genetic, molecular, biochemical, and pharmacological approaches. Many important players of ABA signaling have been identified by genetic screens employing ABA-mediated inhibition of germination or stomatal closure as selection criteria (Koornneef et al., 1984; Merlot et al., 2002). Screening for ABA-insensitive germination thus led to the identification of ABSCISIC ACID-INSENSITIVE1 (ABI1) and ABI2 genes encoding PP2C-type protein phosphatases (Finkelstein and Somerville, 1990), which turned out to be components of the ABA receptor complex (Ma et al., 2009; Park et al., 2009; Klingler et al., 2010). Three of the best characterized positive regulators of ABA signaling are transcription factors encoded by the ABI3, ABI4, and ABI5 genes, which were initially identified by screening for mutants exhibiting ABA-insensitive germination (Koornneef et al., 1984). ABI3, ABI4, and ABI5 belong to B3 DNA binding domain, APETALA2 (AP2), and basic leucine zipper (bZIP) domain protein families, respectively, that regulate overlapping subsets of seed-specific and ABA-inducible genes and control seed maturation and germination (Finkelstein and Somerville, 1990; Giraudat et al., 1992; Parcy and Giraudat, 1997; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Carles et al., 2002; Lopez-Molina et al., 2002; Fujita et al., 2011; Mönke et al., 2012). AtPP2CA was identified in a library of complementary DNA (cDNA)-overexpressing lines, which were screened for the ability to germinate in the presence of inhibitory concentrations of ABA (Kuhn et al., 2006). A similar gain-of-function strategy was employed in the conditional cDNA-overexpressing system (COS) to identify novel components in ABA signal transduction (Papdi et al., 2008). While the ectopic overexpression of regulatory genes may result in dominant dwarfism or reduced fertility (Kasuga et al., 1999; Dinkins et al., 2002), the chemically induced expression in the COS circumvents such disadvantages and permits the generation of fertile transgenic plants (Rigó et al., 2012). Regulated overexpression of cDNAs was employed in the identification of the small heat shock protein gene HSP17.6A, which could confer ABA insensitivity to Arabidopsis (Arabidopsis thaliana), pointing to a novel function of this gene (Papdi et al., 2008).
Here, we report on the functional characterization of the C2H2-type ZINC FINGER PROTEIN3 (ZFP3), which was identified by screening for ABA-insensitive seed germination using the COS. In silico analysis revealed that there are 176 C2H2-type zinc finger proteins in Arabidopsis, from which 33 are conserved in other eukaryotes and 143 appear to be plant specific (Englbrecht et al., 2004). Members of this gene family have been implicated in the regulation of plant development, including photomorphogenesis, leaf, shoot, and flower organogenesis, gametogenesis, seed development, and dormancy (Sakai et al., 1995; Chrispeels et al., 2000; Prigge and Wagner, 2001; Dinkins et al., 2002; He and Gan, 2004; Ohno et al., 2004; Takeda et al., 2004). Our data show that ZFP3 and its closest C2H2-type zinc-finger protein homologs (ZFPs) are negative regulators of ABA signaling during germination, influence vegetative development and fertility, and modulate red light signaling in seedling photomorphogenesis.
RESULTS
ZFP3 Overexpression Confers ABA-Insensitive Seed Germination
Screening for ABA-insensitive seed germination of COS-transformed Arabidopsis plants led to the identification of numerous transgenic lines showing different degrees of ABA insensitivity (Papdi et al., 2008). Germination of line A44 was insensitive to 5 μm ABA in the presence of an estradiol inducer of the chemically regulated promoter in the pER8-GW vector, while it was similar to the wild type in the absence of ABA (Fig. 1, A–C). Germination of A44 was insensitive to paclobutrazol, an inhibitor of GA biosynthesis (Fig. 1D).
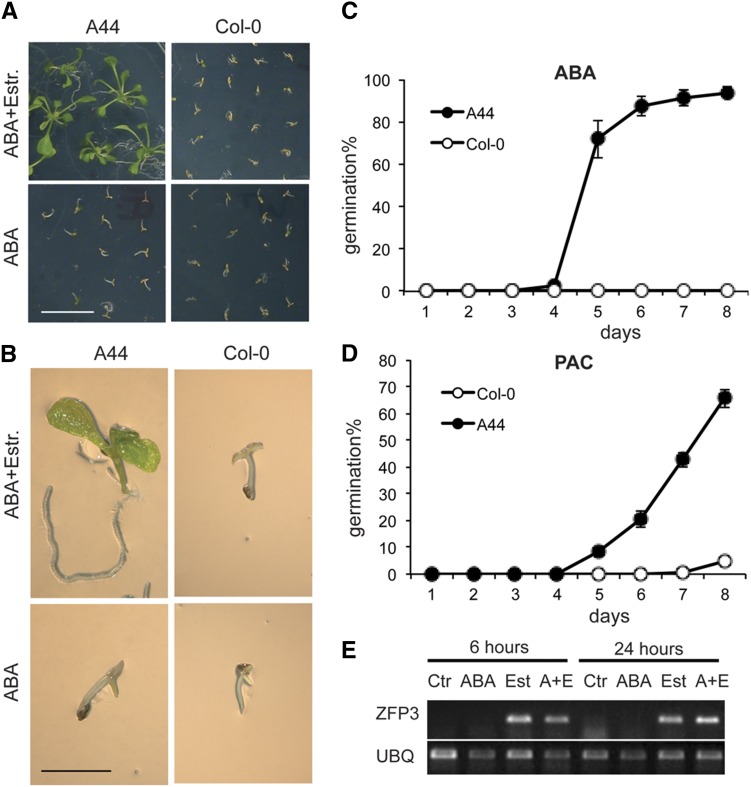
Figure 1.
ABA-insensitive germination of the COS line A44. A, Germination and growth of Col-0 wild-type and A44 seedlings on medium supplemented by 3 μm ABA in the presence or absence of 4 μm estradiol (Estr.). Images show 2-week-old seedlings. Bar = 10 mm. B, Six-day-old Col-0 and A44 seedlings germinating on medium containing 5 μm ABA with or without 4 μm estradiol. Bar = 2 mm. C, Time course of germination in the presence of 5 μm ABA and 4 μm estradiol. D, Germination of Col-0 wild-type and A44 seeds on medium containing 40 μm paclobutrazol (PAC) and 4 μm estradiol. se bars are shown from four repeats. E, Expression of the ZFP3 gene in A44 plants. ZFP3 transcript was detected by reverse transcription-PCR in 2-week-old plantlets treated with ABA and/or estradiol. Ctr, Nontreated control; ABA, 20 μm ABA; Est, 4 μm estradiol; A+E, 20 μm ABA and 4 μm estradiol. The reference gene was UBIQUITIN10 (UBQ; AT4G05320). [See online article for color version of this figure.]
A single PCR fragment was amplified from genomic DNA of A44 plants using ER8A and ER8B primers flanking the cDNA insert (Papdi et al., 2008). The nucleotide sequence of the recovered fragment showed 100% identity with the full-length cDNA of the AT5G25160 gene, encoding ZFP3, the function of which was unknown. Transcription of ZFP3 was not regulated by ABA, and it was only detectable in estradiol-treated A44 plantlets but not in wild-type ones (Fig. 1E). ZFP3 is a member of the large C2H2 zinc finger protein family comprising 176 members in Arabidopsis (Englbrecht et al., 2004). According to phylogenetic classification, ZFP3 belongs to the C1-1iAa subset of C1-1i subfamily (Englbrecht et al., 2004) and is most closely related to ZFP1, ZFP4, ZFP7, and the hypothetical protein encoded by AT5G10970 (Supplemental Fig. S1A). Multiple sequence alignment of the most closely related proteins revealed high similarity between their conserved ZnF_C2H2 domains and C-terminal nuclear localization signals (Supplemental Fig. S1B).
To verify that the ABA insensitivity of A44 was indeed caused by ZFP3 overexpression, we produced transgenic lines expressing the full-length cDNA of ZFP3 fused to coding sequences of a hemagglutinin (HA) epitope tag under the control of an estradiol-inducible promoter (ZFP3ox lines). Western blotting confirmed the presence of the HA-ZFP3 fusion protein in germinating seedlings in the presence of estradiol (Fig. 2A). Germination of seven out of 10 independent ZFP3ox lines was less sensitive to ABA in the presence of estradiol, thereby confirming that the ABA insensitivity of A44 seeds was indeed caused by ZFP3 overexpression (Supplemental Fig. S2).
Figure 2.
ZFP3 modulates ABA sensitivity. A, Western-blot detection of the ZFP3-HA fusion protein in extracts of transformed Arabidopsis seedlings (ZFP3ox) germinated in the light. Protein extracts were prepared from imbibed seeds (day 0) and seedlings germinated for 2 or 3 d in the presence or absence of 2 μm ABA. W., Western blot; C., protein samples stained by Coomassie Brilliant Blue. B, Comparison of the ABA insensitivity of Col-0, ZFP3ox, abi2-1 mutant, and an abi2-1/ZFP3ox line in a germination assay. Germination percentage was scored 8 d after plating imbibed seeds on germination medium containing different concentrations of ABA (0–20 μm). C, Comparison of the ABA insensitivity of Col-0, ZFP3ox, abi4-101 mutant, and an abi4-101/ZFP3ox line in a germination assay. D, Comparison of the ABA insensitivity of Col-0, ZFP3ox, abi5-1 mutant, and an abi5-1/ZFP3ox line in a germination assay. E, Schematic map of T-DNA insertions in zfp3 and zfp4 mutants. Positions of the PCR primers used for monitoring the transcription of mutant alleles are indicated below the maps. F, Germination of zfp3, zfp4, and zfp3 zfp4 on medium supplemented by different concentrations of ABA. Germination rates were scored 7 d after seed plating. G, ABA content of 7-d-old Col-0, ZFP3ox, and zfp3 knockout mutant plants. H, Germination of transgenic Arabidopsis lines overexpressing ZFP1, ZFP3, ZFP4, ZFP6, ZFP7, and ZFP8 cDNAs under the control of an estradiol-inducible XVE promoter in the presence of 2.5 μm ABA with or without 4 μm estradiol. Average germination rates of two independent lines are shown. Error bars indicate se, and different letters show significant differences at P < 0.05 (Duncan’s test).
Next, we compared the ABA responses of ZFP3ox lines and known ABA signaling mutants and found that, in germination assays, the ABA insensitivity of ZFP3ox lines was similar or higher compared with those of the abi2-1, abi4-101, and abi5-1 mutants (Fig. 2, B–D). To analyze the possible genetic interaction of ZFP3 with these ABI genes, the ZFP3ox line was crossed with the abi2-1, abi4-101, and abi5-1 mutants. In germination assays, the abi/ZFP3ox lines showed different levels of enhanced ABA insensitivity when compared with the abi mutants and the ZFP3ox line and were able to germinate even in the presence of 20 μm ABA (Fig. 2, B–D). ZFP3ox thus appeared to increase additively the level of ABA insensitivity conferred by the different abi mutations. These data suggested that ZFP3 is a negative regulator of the ABA-mediated suppression of seed germination and probably acts independently of ABI2, ABI4, and ABI5.
To obtain more precise data on the function of individual ZFP genes, transfer DNA (T-DNA) insertions were identified in ZFP3 and ZFP4 in public mutant collections (Fig. 2E). Quantitative real-time (qRT)-PCR analysis showed that the T-DNA insertion mutations reduced the corresponding transcripts in homozygous zfp3 and zfp4 mutants by at least 1 order of magnitude (Supplemental Fig. S3). The sensitivity of zfp3 and zfp4 mutants to ABA was higher compared with the wild type, whereas the double zfp3 zfp4 mutant displayed more pronounced ABA hypersensitivity, since its germination efficiency was reduced to 40% and 15% of the wild-type level in the presence of 0.4 and 0.6 μm ABA, respectively (Fig. 2F; Supplemental Fig. S3). Overexpression of ZFP3 under the control of an estradiol-inducible promoter in the zfp3 mutant considerably decreased the ABA hypersensitivity of the mutant and reduced the level of ABA sensitivity similar to the wild type or more in most of the tested transgenic lines (Supplemental Fig. S4). To test whether the different germination responses to ABA observed in the ZFP3-overexpressing and mutant lines are due to altered ABA metabolism, the ABA contents of ZFP3ox and the zfp3 mutant were determined. There was no significant change in ABA levels of ZFP3ox and zfp3 plants compared with wild-type plants (Fig. 2G), suggesting that the observed germination phenotype was the result of a change in either ABA perception or signal transduction.
Enhanced ABA sensitivity of the double zfp3 zfp4 mutant suggested that other members of the ZFP gene family might analogously control ABA responses in germination, having at least partially redundant biological functions. Estradiol-dependent overexpression of ZFP1, ZFP4, ZFP6, and ZFP7 in Arabidopsis led to ABA-insensitive germination, comparable to the ZFP3ox line, while enhanced expression of ZFP8 (which is less related to ZFP3 in the phylogenetic tree; Supplemental Fig. S1) had only a minor effect on ABA sensitivity (Fig. 2H).
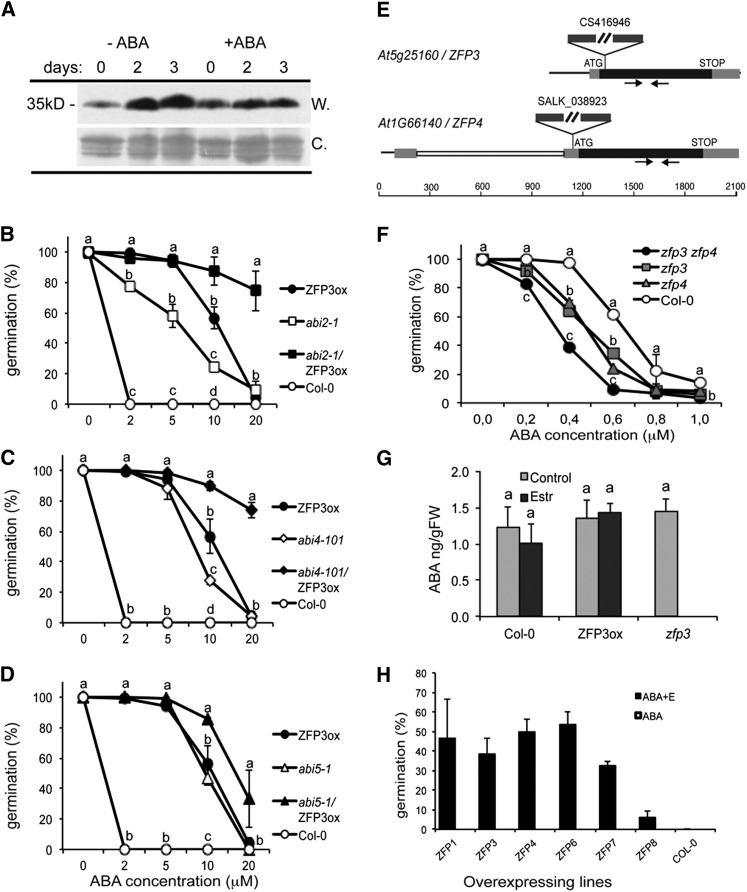
The phenotypes of germinating ZFP3ox or zfp3 mutant seedlings were similar to the wild type. However, overexpression of ZFP3 under the control of the constitutive cauliflower mosaic virus 35S promoter resulted in dwarf plants with considerably reduced fertility. Rosette sizes of ZFP3-overexpressing plants were 50% to 80% smaller than those of wild-type plants (Fig. 3A), while the number and size of the siliques were significantly reduced (Fig. 3, B–D). Most siliques of the ZFP3-overexpressing plants were small and distorted, had few seeds, or were empty (Fig. 3, C–E). Seed yields appeared to inversely correlate with ZFP3 transcript levels in transgenic lines (Supplemental Fig. S5). When compared with the wild type, the germination of 35S-ZFP3 seeds was delayed on one-half-strength Murashige and Skoog culture medium but was insensitive to ABA inhibition (Supplemental Fig. S6). In contrast, the phenotype of the zfp3 mutant was similar to that of the wild type. These pleiotropic effects suggest that ZFP3 not only regulates ABA responses in germination but also plays a role in the control of vegetative and reproductive development.
Figure 3.
Effect of ZFP3 overexpression on plant development. A, Rosette growth of wild-type plants (Col-0) and two independent transgenic plants constitutively expressing ZFP3 (35S-ZFP3). B, Morphology of a flowering 35S-ZFP3 plant. C and D, Siliques of 35S-ZFP3 (C) and wild-type (D) plants. E, Siliques and seeds of 35S-ZFP3 (top silique) and wild-type (bottom silique) plants. Bars = 5 mm. [See online article for color version of this figure.]
Transcriptional Regulation of ZFP3 and Related Genes
Public transcript profiling data (http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi) provided preliminary information about the expression of ZFP3 and its related genes. Transcript profiles of 10 ZFP3-related genes showed considerable development- and organ-specific variations (Supplemental Fig. S7). ZFP3 is expressed in hypocotyl, roots, and to a lesser extent young leaves, shoot apices, and flowers. Expression of ZFP1, ZFP5, ZFP10, and ZFP11 is very low in most organs. ZFP2 transcription is high in flowers and developing seeds, ZFP6 expression is high in hypocotyls, roots, and stems, while ZFP4, ZFP7, and ZFP8 transcripts were detected mostly in leaves, roots, cotyledons, and shoot apices. The responses to plant hormones of these ZFP genes also varied considerably. While ABA reduced the transcript levels of most ZFP genes, including ZFP3, it enhanced the expression of ZFP5 (Supplemental Fig. S8).
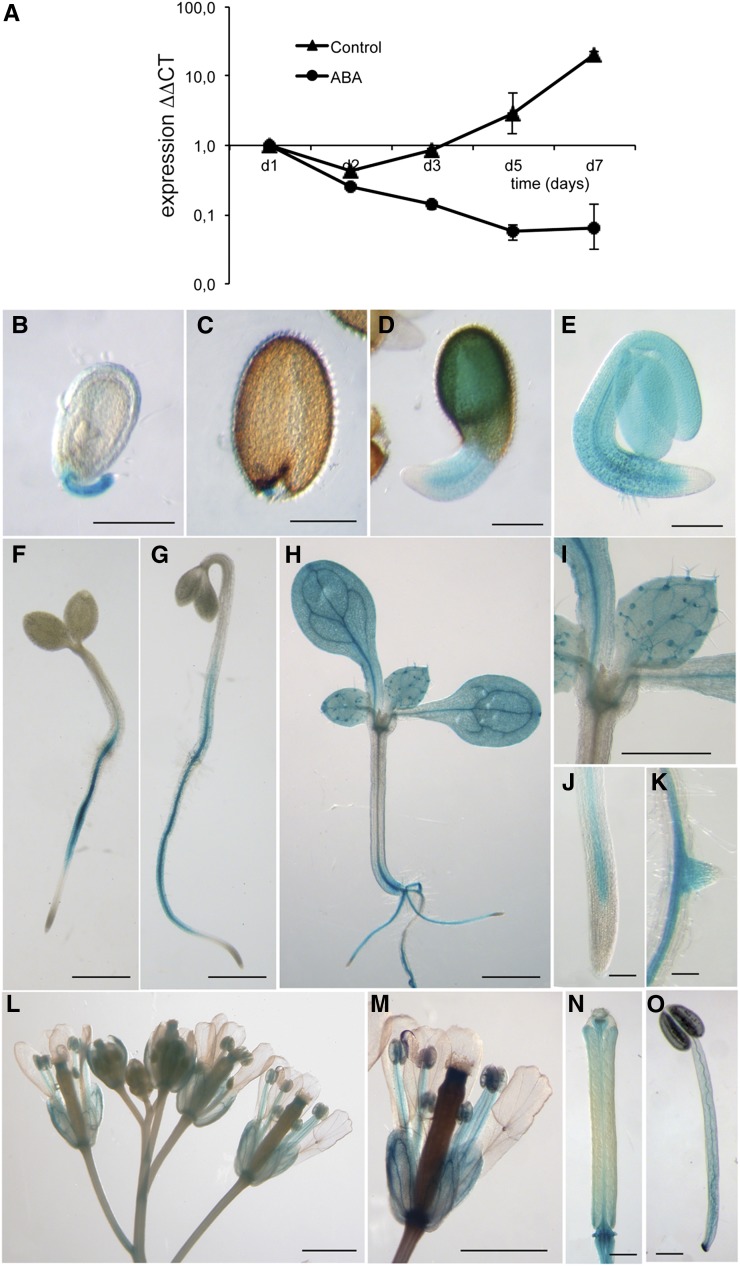
To test the expression of ZFP3 during germination, transcript levels were monitored in imbibed seeds and young seedlings. After a slight decline, ZFP3 transcript levels increased steadily during germination. When seeds were germinated on ABA-containing medium, ZFP3 transcription was reduced (Fig. 4A), correlating with the public transcript data.
Figure 4.
Transcriptional regulation of the ZFP3 gene. A, qRT-PCR analysis of ZFP3 expression during germination. Wild-type seeds were germinated in the absence (Control) or presence of 2 μm ABA (ABA). GAPC2 (At1g13440) was used as a reference transcript. Relative transcript values are shown, where 1 equals transcript levels at the start of germination (Fohgrub and Kempken, 2012). ΔΔCT, Delta delta cycle threshold. B to O, Detection of pZFP3-GUS activity in transgenic Arabidopsis. Histochemical localization of GUS activity is shown in developing and mature seeds (B and C), germinating seeds (D and E), 3-d-old seedlings germinated in light (F) or dark (G), 7-d-old plantlets (H–K), and flowers (L–O) of transgenic plants. Bars = 0.1 mm (B–E, J, K, N, and O) and 1 mm (F–I, L, and M).
Spatial transcriptional regulation of ZFP3 expression was investigated by testing the activity of the ZFP3 promoter-GUS reporter gene construct (pZFP3-GUS) in transgenic Arabidopsis plants using 5-bromo-4-chloro-3-indolyl-β-glucuronic acid histochemical staining. GUS activity was present in the chalazal/micropylar axis in developing and mature seeds but not in the embryo itself (Fig. 4, B and C), and it could be detected in emerging radicles and later in hypocotyls, cotyledons, and radicles of germinating seedlings (Fig. 4, D and E). In young seedlings, pZFP3-GUS activity was present in roots and hypocotyls of seedlings (Fig. 4, F and G). In older plants, GUS was detectable in leaves and roots, in particular in emerging lateral roots (Fig. 4, H–K). GUS activity was present in flowers, in particular in sepals and stamens (Fig. 4, L–O). These data suggest that ZFP3 is expressed in most organs, showing high expression in vascular tissues.
ZFP3 Is a Nuclear Protein
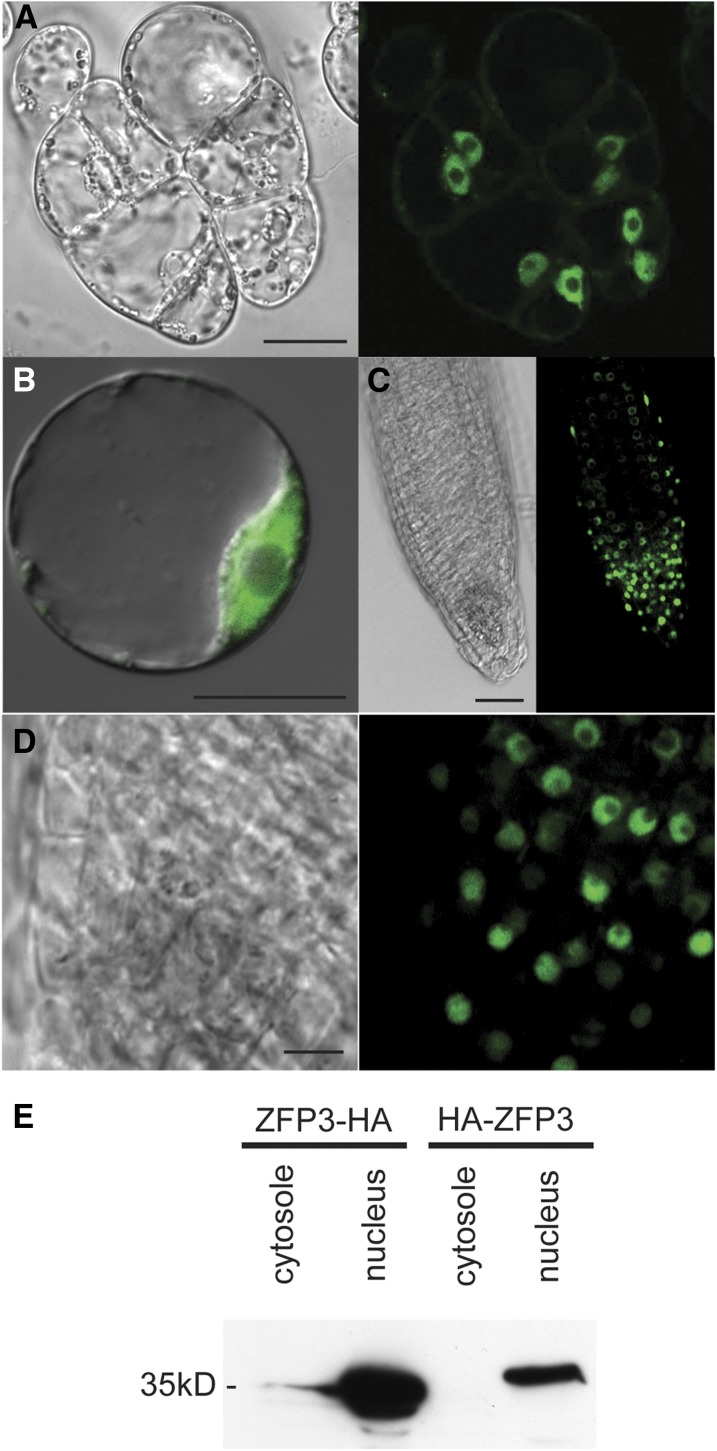
Nuclear localization signals were found in ZFP3 and related ZFP proteins (Supplemental Fig. S1). To verify its intracellular localization, ZFP3 was fused to an enhanced GFP (eGFP) marker and expressed in cultured cells and transgenic plants under the control of the cauliflower mosaic virus 35S promoter. The fluorescence pattern of eGFP suggested that the ZFP3 protein is localized in nuclei but not in the nucleolus (Fig. 5, A–D). To confirm these data, both N- and C-terminal HA-tagged ZFP3 fusion proteins were expressed in Arabidopsis under the control of the 35S promoter, and the abundance of ZFP3-HA was determined in subcellular fractions separated by differential centrifugation. Western blotting detected both HA-ZFP3 and ZFP3-HA only in nuclear but not in other cellular fractions, thereby confirming the nuclear localization of the ZFP3 protein (Fig. 5E).
Figure 5.
Intracellular localization of the ZFP3 protein. A and B, Nuclear localization of the ZFP3-eGFP fusion protein in transformed cultured cells (A) and protoplasts (B). Bars = 20 μm. C and D, ZFP3-eGFP is localized in nuclei of root cells of transgenic plants. Bars = 50 μm. E, Detection of ZFP3-HA and HA-ZFP3 fusion proteins in nuclear and cytosolic protein fractions prepared from transformed Arabidopsis cells. Bar = 20 μm.
ZFP3 Modulates ABA-Regulated Gene Expression
Although the majority of known C2H2_Zn finger proteins are transcriptional regulators, they can also be involved in other conserved biological processes, such as chromatin remodeling, RNA binding, or protein-protein interactions (Riechmann et al., 2000; Englbrecht et al., 2004). To identify potential ZFP3-regulated genes, whole-genome transcript profiling was performed with ZFP3ox and Columbia-0 (Col-0) seedlings germinated in the presence of 5 μm estradiol with or without 2.5 μm ABA. Seedlings with similar developmental stages, having emerged radicles but not open cotyledons, were used for transcript analysis. As knockout mutation leads to only a weak ABA-sensitive phenotype, the zfp3 mutant was not involved in this study. Transcript levels were determined by in-depth RNA sequencing, and normalized expression values were compared. Genes with fewer than 30 reads were excluded from analysis; therefore, the expression of 23,932 genes was analyzed. ZFP3 overexpression resulted in up- and down-regulated expression of 222 and 153 genes, respectively (Supplemental Data Set S1). According to the Genevestigator database, many genes that are up- or down-regulated by ZFP3 are repressed or induced during germination, respectively (Supplemental Data Sets S2 and S3; Supplemental Figs. S9–S11). ZFP3 overexpression considerably modified the expression profiles of many ABA-regulated genes. While in our experimental conditions, ABA enhanced the expression of 802 genes, transcript levels of 27% of these genes (217) were reduced by at least 2-fold in ZFP3ox seedlings (Table I; Supplemental Data Set S1). According to Gene Ontology classification, proteins that are involved in developmental regulation, such as embryo and seed development, response to desiccation, water deprivation, and ABA stimulus, and localized in nuclei and mitochondria were overrepresented in this category (Table I; Supplemental Fig. S9). Numerous genes in this group were induced by ABA, drought, and salt stress, up-regulated in cop9 signalosome subunit3-1 (csn3-1), csn4-1, csn5, the quadruple heat shock factor (hsf)a1a, hsfa1b, hsfa1d, hsfa1e, and the quintuple phytochrome (phy)ABCDE mutants, and repressed during germination, by white and red light, and in greening after extended darkness1 (ged1) and Phy-interacting factor1 (pif1), pif3, pif4, pif5 quadruple mutants (Supplemental Data Set S4; Supplemental Fig. S12). Transcript levels of 534 genes were reduced in seedlings on ABA-containing medium, while 118 genes in this category (22%) were less inhibited by ABA in ZFP3ox. Many of these genes are involved in photosynthesis and light responses (Table II; Supplemental Data Set S5; Supplemental Fig. S13). Gene Ontology terms of electron transport, energy and cell organization, and plastid localization were overrepresented in this category (Supplemental Fig. S9). Most of the ABA-suppressed and ZFP3-derepressed genes are down-regulated by stress conditions such as drought, pathogen infection, and the elicitor 22-amino acid sequence of flagellin (FLG22) and in brevis radix (brx), Abscisic Acid-hypersensitive germination1 (ahg1-1), ahg3-1, csn3-1, csn4-1, csn5, and the quintuple phyABCDE mutants. Many genes in this category are activated during germination and shoot regeneration and by light and have enhanced expression in phytoalexin deficient4 (pad4), enhanced disease susceptibility1-2 (eds1-2), and pif1,3,4,5 quadruple mutants (Table III; Supplemental Data Set S5; Supplemental Fig. S13). These results suggest that a subset of genes that are inversely regulated by ABA and ZFP3 are also controlled by light through a phytochrome light receptor and one or more PIF transcription factors.
Table I. Thirty-three most ABA-induced genes.
Thirty-one of these genes are less induced in ZFP3ox than in the Col-0 background.
| Gene | ABA (Col-0) | ABA (ZFPox) | ZFPox (Control) | ZFPox (ABA) | Arabidopsis Genome Initiative Code | Protein |
|---|---|---|---|---|---|---|
| RAB18 | 45.8 | 16.4 | −1.3 | −3.5 | AT5G66400 | Responsive to ABA18 (RAB18) |
| AT3G15670 | 42.8 | 15.9 | −1.5 | −4.1 | AT3G15670 | Late embryogenesis abundant protein (LEA) |
| AT3G54940 | 38.9 | 4.9 | 1.5 | −5.3 | AT3G54940 | Papain family Cys protease |
| OLEO2 | 34.4 | 18.6 | 2.1 | 1.1 | AT5G40420 | Oleosin2 (OLEO2) |
| AT3G02480 | 33.7 | 30.2 | −2.0 | −2.2 | AT3G02480 | Late embryogenesis abundant protein (LEA) |
| AT3G01570 | 31.8 | 7.2 | 1.8 | −2.4 | AT3G01570 | Oleosin family protein |
| AT3G17520 | 30.6 | 12.4 | −1.4 | −3.5 | AT3G17520 | Late embryogenesis abundant protein (LEA) |
| LEA4-5 | 27.5 | 8.6 | −1.0 | −3.3 | AT5G06760 | Late embryogenesis abundant4-5 (LEA4-5) |
| GEA6 | 27.2 | 10.9 | −1.2 | −3.0 | AT2G40170 | Late embryogenesis abundant6 (GEA6) |
| AT4G36600 | 26.0 | 5.8 | 1.4 | −3.3 | AT4G36600 | Late embryogenesis abundant protein (LEA) |
| AT2G28490 | 26.0 | 7.2 | 1.0 | −3.6 | AT2G28490 | RmlC-like cupins superfamily protein |
| AT5G35660 | 25.4 | 10.7 | −1.5 | −3.6 | AT5G35660 | Gly-rich protein |
| AT1G04560 | 23.8 | 12.5 | −2.1 | −3.9 | AT1G04560 | AWPM-19-like family protein |
| EM1 | 23.6 | 6.0 | 1.5 | −2.6 | AT3G51810 | Late embryogenesis abundant1 (EM1) |
| OLEO1 | 22.1 | 7.8 | −1.2 | −3.4 | AT4G25140 | Oleosin1 (OLEO1) |
| LEA18 | 21.9 | 11.7 | −1.2 | −2.3 | AT2G35300 | Late embryogenesis abundant18 (LEA18) |
| PER1 | 20.8 | 5.8 | 1.5 | −2.3 | AT1G48130 | 1-Cys peroxiredoxin1 (PER1) |
| LEA7 | 20.6 | 9.1 | −1.5 | −3.5 | AT1G52690 | Late embryogenesis abundant protein (LEA) |
| AT5G51370 | 20.3 | 1.4 | 1.1 | −13.0 | AT5G51370 | RNI-like superfamily protein |
| PAP85 | 19.5 | 2.5 | 1.3 | −6.2 | AT3G22640 | Nutrient reservoir protein (PAP85) |
| AT3G52260 | 19.3 | 4.3 | 1.2 | −3.6 | AT3G52260 | Pseudouridine synthase family protein |
| AT3G50520 | 19.1 | 1.4 | 8.9 | −1.5 | AT3G50520 | Phosphoglycerate mutase family protein |
| LEA | 18.8 | 4.2 | −1.0 | −4.6 | AT2G21490 | Dehydrin late embryogenesis abundant protein (LEA) |
| AT2G15010 | 18.1 | 3.0 | 2.0 | −3.0 | AT2G15010 | Pathogenesis-related protein (PR) |
| GASA3 | 17.4 | 8.1 | 1.1 | −2.0 | AT4G09600 | GAST1 protein homolog3 (GASA3) |
| AT5G66780 | 17.1 | 6.0 | −1.2 | −3.5 | AT5G66780 | Unknown protein |
| TIP3;1 | 15.9 | 6.9 | 1.1 | −2.2 | AT1G73190 | α-Tonoplast intrinsic protein (TIP3;1) |
| CRA1 | 14.9 | 5.4 | −1.4 | −3.9 | AT5G44120 | Cruciferina (CRA1) |
| AT2G25890 | 14.1 | 5.5 | 1.1 | −2.4 | AT2G25890 | Oleosin family protein |
| AT3G03341 | 14.0 | 6.5 | −1.4 | −3.0 | AT3G03341 | Unknown protein |
| AT1G54870 | 13.9 | 5.7 | −1.1 | −2.7 | AT1G54870 | ChlADR aldehyde reductase |
| LTI65 | 13.4 | 8.6 | −2.0 | −3.1 | AT5G52300 | Low-temperature-induced65 (LTI65) |
| HVA22B | 13.3 | 4.0 | −1.1 | −3.6 | AT5G62490 | HVA22 homolog B (HVA22B) |
Table II. Thirty-three genes most down-regulated by ABA.
Twenty-eight of these genes are less suppressed in ZFP3ox seedlings.
| Gene | ABA (Col-0) | ABA (ZFPox) | ZFPox (Control) | ZFPox (ABA) | Arabidopsis Genome Initiative Code | Protein |
|---|---|---|---|---|---|---|
| LHCB2.1 | −39.1 | −1.5 | −1.2 | 21.5 | AT2G05100 | PSII light-harvesting complex2.1 (LHCB2.1) |
| CAB1 | −29.8 | −2.4 | 1.5 | 19.0 | AT1G29930 | Chlorophyll a/b-binding protein1 (CAB1) |
| LHB1B2 | −25.8 | −1.6 | −1.2 | 13.9 | AT2G34420 | PSII light-harvesting complex B1B2 (LHB1B2) |
| LHCB2.2 | −25.1 | −1.5 | −1.2 | 13.5 | AT2G05070 | PSII light-harvesting complex2.2 (LHCB2.2) |
| AT4G23680 | −25.0 | −4.6 | −1.6 | 3.5 | AT4G23680 | Polyketide cyclase/dehydrase and lipid transport protein |
| LHCB3 | −19.4 | −2.1 | −1.1 | 8.2 | AT5G54270 | Light-harvesting chlorophyll b-binding protein3 (LHCB3) |
| PSBO1 | −19.3 | −2.5 | 1.2 | 9.1 | AT5G66570 | PSII oxygen-evolving complex1 (PSBO1) |
| PORB | −18.6 | −8.3 | −1.3 | 1.8 | AT4G27440 | Protochlorophyllide oxidoreductase B (PORB) |
| LHCB5 | −18.1 | −2.3 | 1.2 | 9.9 | AT4G10340 | PSII light-harvesting complex5 (LHCB5) |
| PSAH2 | −17.5 | −3.5 | 1.0 | 5.0 | AT1G52230 | PSI subunit H2 (PSAH2) |
| DOT1 | −17.3 | −2.5 | −3.3 | 2.2 | AT2G36120 | Defectively organized tributaries1 (DOT1) |
| RBCS3B | −16.6 | −5.2 | −1.3 | 2.4 | AT5G38410 | Rubisco small subunit 3B (RBCS3B) |
| AT3G48140 | −15.6 | −5.0 | −2.0 | 1.5 | AT3G48140 | B12D protein |
| RCA | −15.3 | −3.7 | −1.3 | 3.3 | AT2G39730 | Rubisco activase (RCA) |
| CAB2 | −14.8 | −2.0 | 1.6 | 11.8 | AT1G29920 | Chlorophyll a/b-binding protein2 (CAB2) |
| CCoAOMT1 | −14.6 | −3.2 | −1.9 | 2.3 | AT4G34050 | Caffeoyl-CoA o-methyltransferase1 (CCoAOMT1) |
| LHCA1 | −14.5 | −1.5 | 1.1 | 10.4 | AT3G54890 | PSI light-harvesting complex1 (LHCA1) |
| GAMMA-TIP | −13.7 | −3.1 | −1.5 | 2.9 | AT2G36830 | γ-Tonoplast intrinsic protein (GAMMA-TIP) |
| LHCB6 | −13.2 | −3.0 | 1.5 | 6.5 | AT1G15820 | PSII light-harvesting complex6 (LHCB6) |
| AT1G09310 | −13.2 | −3.9 | −1.3 | 2.6 | AT1G09310 | Protein of unknown function |
| CAD9 | −13.1 | −3.9 | −1.7 | 1.9 | AT4G39330 | Cinnamyl alcohol dehydrogenase9 (CAD9) |
| AT1G29090 | −13.0 | −4.9 | −2.8 | −1.0 | AT1G29090 | Cys proteinases superfamily protein |
| ACO2 | −12.8 | −3.5 | −1.7 | 2.2 | AT1G62380 | ACC oxidase2 (ACO2) |
| LHCA2 | −12.4 | −2.3 | 1.3 | 6.9 | AT3G61470 | PSI light-harvesting complex 2 (LHCA2) |
| PSAL | −12.0 | −2.4 | 1.1 | 5.5 | AT4G12800 | PSI subunit L (PSAL) |
| PSAN | −11.9 | −2.0 | 1.0 | 6.0 | AT5G64040 | Subunit of PSI (PSAN) |
| CAB3 | −11.5 | −1.5 | 1.2 | 9.6 | AT1G29910 | Chlorophyll a/b-binding protein3 (CAB3) |
| TIP2 | −11.4 | −3.7 | −1.2 | 2.5 | AT3G26520 | Tonoplast intrinsic protein2 (TIP2) |
| PSAO | −11.2 | −2.0 | 1.2 | 6.7 | AT1G08380 | PSI subunit O (PSAO) |
| LHCA3 | −10.7 | −2.3 | 1.0 | 4.7 | AT1G61520 | PSI light-harvesting complex3 (LHCA3) |
| PIP1A | −10.6 | −2.2 | −1.7 | 2.8 | AT3G61430 | Plasma membrane intrinsic protein1A (PIP1A) |
| LHCB4.1 | −10.6 | −2.2 | 1.1 | 5.6 | AT5G01530 | PSII light-harvesting complex (LHCB4.1) |
| AT5G17820 | −10.4 | −4.1 | −2.5 | 1.0 | AT5G17820 | Peroxidase superfamily protein |
Table III. Expression profiles of ABA- and ZFP3-regulated genes in the Genevestigator transcript profiling database.
Four groups of genes were analyzed with the hierarchical clustering tool of the Genevestigator database: those that were down- or up-regulated in ZFP3ox line, induced by ABA but down-regulated by ZFP3ox, and repressed by ABA but up-regulated by ZFP3ox (Supplemental Data Set S1). Percentage values indicate the proportions of genes similarly regulated by a given treatment or in a mutant. Hierarchical clustering data are given in Supplemental Data Sets S2 to S5. Visualized data of hierarchical clustering are shown in Supplemental Figures S10 to S13. Descriptions of the listed mutants are available in Supplemental Text S1.
| Categories | Genes Down-Regulated by ZFP3 | Genes Up-Regulated by ZFP3 | Genes Induced by ABA but Down-Regulated by ZFP3 | Genes Repressed by ABA but Up-Regulated by ZFP3 |
|---|---|---|---|---|
| Induced by (treatments, conditions) | Germination (32%) | 5-Azacytidine (17%) | ABA (57%) | Germination (84%) |
| Phytophthora parasitica (12%) | Drought (33%) | Shoot regeneration (60%) | ||
| Osmotic stress (33%) | White light (45%) | |||
| Salt stress (26%) | Red light (40%) | |||
| Repressed by (treatments, conditions) | 5-Azacytidine (24%) | Germination (13%) | Germination (62%) | ABA (81%) |
| Salt (root protoplasts 21%) | Red light (37%) | 5-AC (67%) | ||
| White light (34%) | Paclobutrazol (64%) | |||
| Drought (54%) | ||||
| Callus (59%) | ||||
| Hypoxia (59%) | ||||
| Pseudomonas syringae (59%) | ||||
| FLG22 (50%) | ||||
| Norflurazon (48%) | ||||
| Induced in (mutants, lines) | brx (22%) | hsfa1a,1b,1d,1e (50%) | pif1,3,4,5 (52%) | |
| LEAFY COTYLEDON1 overexpression (46%) | pad4 (46%) | |||
| 35S::amiR(MIR172a) (46%) | eds1-2 (36%) | |||
| phyABCDE (42%) | ||||
| csn4-1 (30%) | ||||
| csn3-1 (25%) | ||||
| csn5 (25%) | ||||
| Repressed in (mutants, lines) | timing of cab expression1 (12%) | ged1 (52%) | csn3-1 (66%) | |
| caprice-1, triptychon-82 (11%) | pif1,3,4,5 (37%) | csn4-1 (67%) | ||
| MAP Kinase6_1 (34%) | csn5 (65%) | |||
| brx (57%) | ||||
| ahg1-1 (59%) | ||||
| ahg3-1 (50%) | ||||
| 35S::amiR(MIR172a) (47%) | ||||
| phyABCDE (40%) |
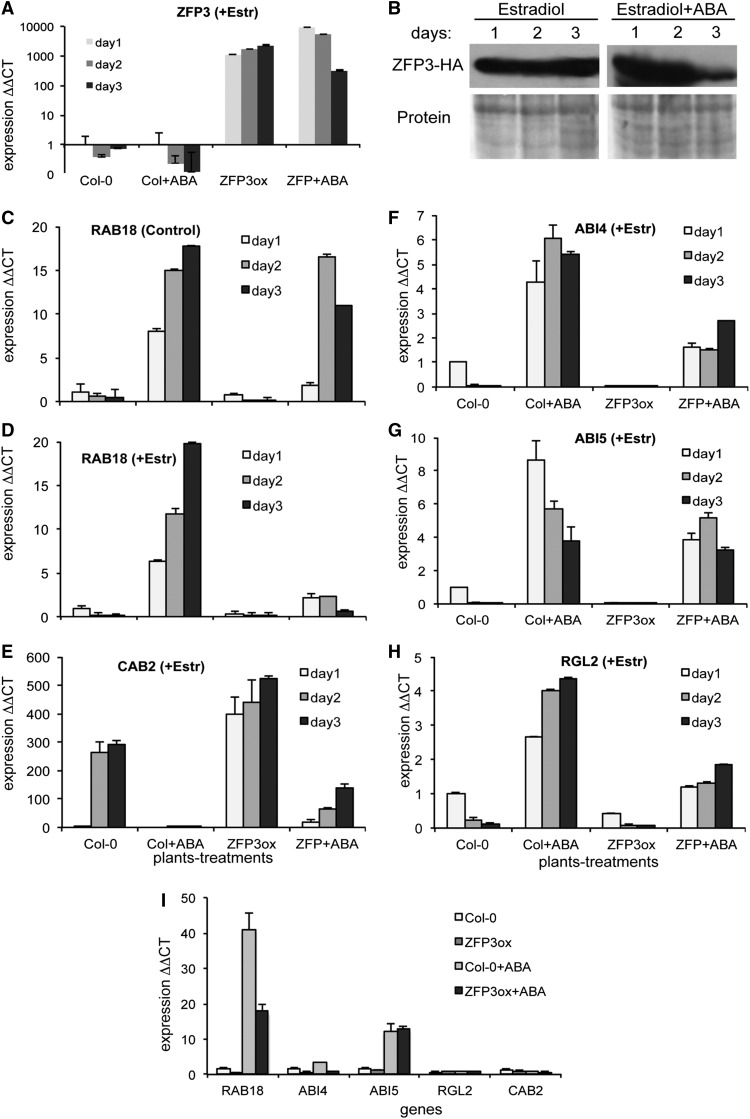
qRT-PCR analysis was employed to investigate further the expression profiles of selected genes in wild-type and ZFP3ox seedlings. In this experiment, enhanced transcription of ZFP3 was confirmed by qRT-PCR and the presence of the HA-tagged ZFP3 was detected using western blotting (Fig. 6, A and B). ABA-dependent activation of the dehydrin gene RESPONSIVE TO ABSCISIC ACID18 (RAB18) was characteristic in wild-type seedlings but was considerably lower in ZFP3ox seedlings in the presence of estradiol (Fig. 6, C and D). Expression of the light-induced gene CHLOROPHYLL A/B-BINDING PROTEIN2 (CAB2) was reduced in wild-type seedlings in the presence of ABA, which was relieved in ZFP3ox seedlings (Fig. 6E). The transcription factors ABI4 and ABI5 are positive regulators of ABA-responsive genes, while RGA-LIKE2 (RGL2) is a repressor of GA signal transduction. Transcript levels of these genes were enhanced by ABA in wild-type seedlings, which was 2 to 4 times lower in ZFP3ox plantlets (Fig. 6, F–H). To compare short-term activation of the tested genes in ZFP3ox and Col-0 plants, 3-d-old seedlings were treated with ABA in the presence of estradiol and transcript levels were tested after 3 h. ABA-dependent transcriptional activation of RAB18 and ABI4 was lower in ZFP3ox plantlets, while the expression of CAB2, ABI5, and RGL2 was not influenced by ZFP3 in these conditions (Fig. 6I). These results showed that, in young plants, ZFP3 can interfere with ABA signals, modulating the activation of a subset of genes. Other genes were probably not direct targets of ZFP3, and differences in their transcript levels were due to developmental effects or controlled by other ZFP3-regulated factors.
Figure 6.
ZFP3 modulates ABA-controlled gene expression. A to H, Seedlings were germinated on 4 μm estradiol-containing medium in the absence or presence of 2 μm ABA, and gene expression was monitored in 1-, 2-, and 3-d-old seedlings. A, Expression of ZFP3 in wild-type and ZFP3ox seedlings. B, Western-blot detection of HA-tagged ZFP3 in overexpressing seedlings. C and D, Transcript levels of RAB18 in germinating seedlings in the absence (C) or presence (D) of estradiol. E, Transcript levels of the light-induced CAB2 gene, encoding a chlorophyll a/b-binding protein, in germinating seedlings treated with estradiol. F to H, Transcript levels of ABI4 and ABI5 (F and G) and RGL2 (H) in germinating seedlings treated with estradiol. I, Expression of the above-tested genes in 3-d-old wild-type (Col-0) and ZFP3ox plantlets induced by 24 μm ABA for 3 h. All plants were treated with 4 μm estradiol for 24 h prior to ABA induction. Expression was normalized to transcript levels at the start of ABA treatment. Relative transcript values are shown; 1 corresponds to transcript levels at the start of experiment (Fohgrub and Kempken, 2012). ΔΔCT, Delta delta cycle threshold. Error bars show se. GAPC2 (At1g13440) was used as constitutive reference.
ZFP3 Modulates Light Responses in Seedling Development
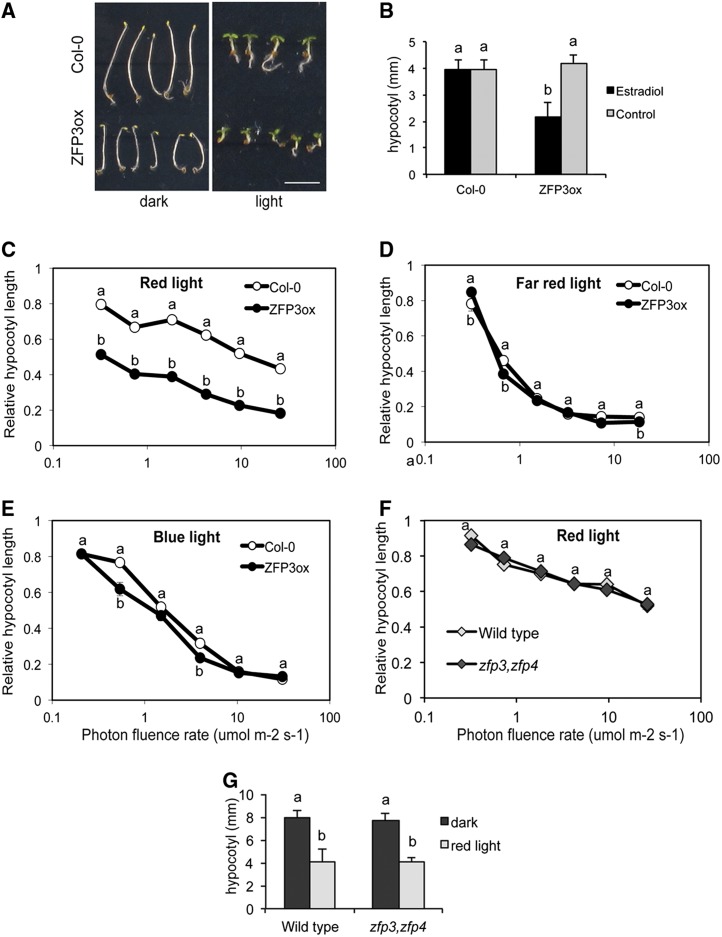
Transcript levels of many light-regulated genes were altered in ABA-treated ZFP3ox seedlings, suggesting that ZFP3 overexpression could influence light signaling. Besides regulating the expression of photosynthetic genes, light controls photomorphogenesis and inhibits hypocotyl elongation. Although interactions between different hormones and light signaling have been studied extensively (Chen et al., 2008; Seo et al., 2009; Lau and Deng, 2010; Hsieh et al., 2012), our understanding of the underlying complex regulation is still far from complete. Visual inspection of young seedlings indicated that, in the presence of estradiol, ZFP3ox plants produced shorter hypocotyls than wild-type plants, both in the dark and in white light (Fig. 7A). This response was estradiol dependent, indicating that it is the result of ZFP3 overexpression (Fig. 7B). Estradiol-dependent shortening of hypocotyls was also observed in transgenic seedlings overexpressing ZFP1, ZFP4, ZFP6, and ZFP7 under the control of the XVE expression system (Supplemental Fig. S14; Zuo et al., 2000). The short-hypocotyl phenotype of ZFP3ox and ZFP1ox seedlings could not be reversed with exogenously added GA or epibrassinolide, hormones known to promote cell elongation. In the dark, the hypocotyl elongation of Col-0 seedlings was reduced by 50% on 1-aminocyclopropane-1-carboxylic acid (ACC)-containing plates, while hypocotyls of ZFP3ox and ZFP1ox seedlings were only 10% shorter (Supplemental Fig. S15). The apical hook of dark-germinated ZFP3ox and ZFP1ox seedlings was similar to that of Col-0 on ACC-supplemented and control plates, respectively (data not shown). The germination of ZFP3ox and zfp3 zfp4 double mutant seeds, however, was not influenced by ACC or epibrassinolide (Supplemental Fig. S16).
Figure 7.
ZFP3 modulates light-controlled hypocotyl elongation. A, Three-day-old Col-0 wild-type and ZFP3-overexpressing seedlings grown in the dark and light. Seeds were germinated in the presence of 4 μm estradiol. Bar = 5 mm. B, Change of hypocotyl lengths in white light in the absence or presence of estradiol. C to E, Relative hypocotyl lengths of wild-type and ZFP3ox seedlings grown under different fluence rates of monochromatic red (C), far-red (D), and blue (E) light. F, Relative hypocotyl lengths of wild-type and zfp3 zfp4 double mutant seedlings germinated under different intensities of red light. G, Hypocotyl lengths of wild-type and zfp3 zfp4 double mutant seedlings grown either in the dark or in red light at a 35 µmol m−2 s−1 fluence rate. Error bars indicate se values. Different letters show significant differences at P < 0.05 (Duncan’s test [B and G] or Student’s t test [C–F]). In C to F, hypocotyl lengths were normalized to the corresponding dark-grown seedlings, and the x axes show a logarithmic scale. [See online article for color version of this figure.]
Visible light perception is mainly mediated by photoreceptors, including phytochromes absorbing red/far-red light and phototropins and cryptochromes sensing blue light (Franklin and Quail, 2010; Chaves et al., 2011). To identify the particular light signaling pathway that is modulated by ZFP3, hypocotyl elongation was tested under different intensities of monochromatic light of different wavelengths. In order to reveal light-dependent effects of ZFP3ox, hypocotyl lengths were normalized to the average length of the corresponding dark-grown seedlings. Relative hypocotyl lengths of ZFP3ox seedlings were reduced in red light (Fig. 7C) but were similar to the wild type under far-red or blue light (Fig. 7, D and E). In contrast, hypocotyl lengths of zfp3 zfp4 double mutants were identical to that of the wild type in the dark or under different fluence rates of red light (Fig. 7, F and G). These data suggest that ZFP3 is implicated in red light-induced responses and acts additively with the function of a red light receptor but does not interfere with far-red or blue light signaling. The fact that overexpression of other ZFP genes reduced hypocotyl elongation, which was not altered in single or double zfp3 zfp4 mutants, suggests that members of this gene subfamily have overlapping functions in the control of photomorphogenesis.
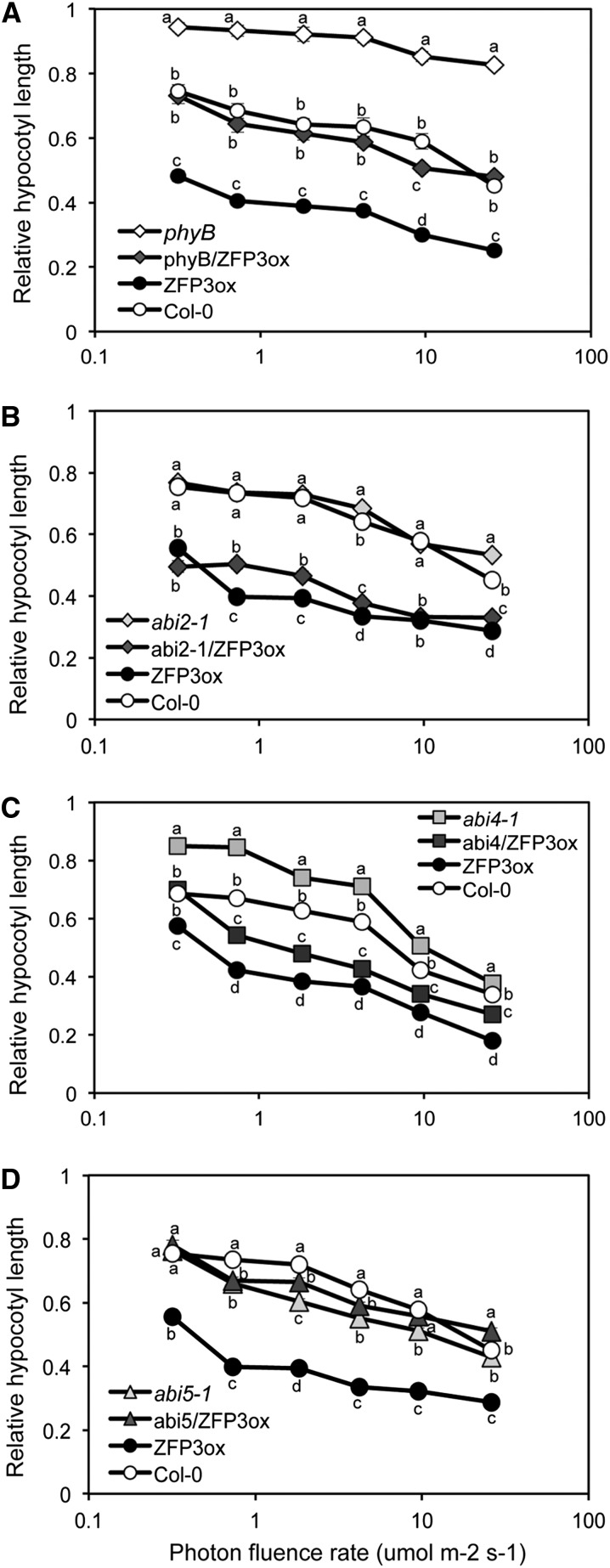
The red/far-red light-absorbing phytochromes are encoded by a small multigene family in Arabidopsis. PhyB and PhyA are considered the major receptors of red and far-red light signals, respectively (Franklin and Quail, 2010). The fact that ZFP3ox showed no altered hypocotyl elongation in far-red or blue light strongly suggested that ZFP3 does not function in PhyA- or cryptochrome-dependent signal transduction. To investigate possible genetic interactions of ZFP3 with PhyB, the ZFP3ox line was crossed with the phyB mutant and the hypocotyl elongation of phyB, ZFP3ox, and phyB/ZFP3ox seedlings was compared at different fluence rates of red light. Hypocotyl lengths of phyB were longer than those of wild-type and ZFP3ox seedlings and were barely changed by different red light intensities, while ZFP3ox reduced hypocotyl lengths in both Col-0 and phyB backgrounds (Fig. 8A). Importantly, hypocotyls of phyB/ZFP3ox seedlings were gradually shortened by increasing fluence rates of red light, similar to wild-type seedlings (Fig. 8A). These results collectively indicate that ZFP3 is implicated in red light signaling independent of PhyB and suggest a role for ZFP3 in the amplification of red light signals. In germination tests, we showed that ZFP3 overexpression increased the ABA insensitivity of abi2-1, abi4-101, and abi5-1 mutants (Fig. 2, B–D). To investigate the light responses of these ABA signaling mutants and to test if they are influenced by ZFP3, hypocotyls of abi/ZFP3ox lines were measured in the light and dark. Hypocotyl lengths in the dark were reduced by ZFP3ox in abi2-1 and abi5-1, while in abi4-101, whose hypocotyls were 30% shorter than those in the wild type, hypocotyl elongation was barely changed by ZFP3 overexpression. In white light, ZFP3 overexpression in the Col-0, abi2-1, and abi4-101 backgrounds reduced hypocotyl lengths by 50% to 60%, while in abi5-1/ZFP3ox seedlings, hypocotyls were only 10% shorter than in abi5-1 (Supplemental Fig. S17). The effect of red light on relative hypocotyl lengths (normalized to hypocotyls of dark-germinated seedlings) was comparable in abi2-1 and Col-0 seedlings, which were reduced to similar extents by ZFP3 overexpression (Fig. 8B). In contrast, the inhibition of hypocotyl elongation in abi4-101 was less sensitive to red light, especially at lower fluence rates, whereas the abi4/ZFP3ox seedlings had significantly shorter hypocotyls than abi4-101 but longer ones when compared with ZFP3ox seedlings (Fig. 8C). The relative hypocotyl lengths of Col-0, abi5-1, and abi5/ZFP3ox seedlings were similar at different fluence rates of red light. In contrast to the Col-0 background, ZFP3 overexpression could not reduce relative hypocotyl lengths in the abi5-1 mutant background upon red light illumination (Fig. 8D). The fact that abi5-1 prevents ZFP3-promoted hypocotyl shortening suggests that ABI5 regulates this photomorphogenic response downstream of ZFP3 and is essential for the light-dependent function of ZFP3.
Figure 8.
Genetic interaction of ZFP3 with PhyB and ABA signaling components. Relative hypocotyl lengths of seedlings with the indicated genotypes are shown at different fluence rates of red light in the presence of 4 μm estradiol. Hypocotyl lengths were normalized to the corresponding dark-grown seedlings. The x axes show a logarithmic scale. Statistical tests were performed by one-way ANOVA and Tukey’s test independently for each fluence rate, and different letters show significant differences at P < 0.05.
DISCUSSION
The phytohormones GA and ABA are major, but not the only, regulators of seed germination, which is promoted by GA and inhibited by ABA. We employed ABA-repressed seed germination as the selection criterion to identify cDNA-overexpressing lines showing ABA insensitivity. Line A44 displayed clear estradiol-dependent ABA insensitivity, suggesting that regulated overexpression of the inserted ZFP3 cDNA is responsible for the observed phenotype (Fig. 1). The causal relationship between ZFP3 overexpression and the ABA-insensitive seed germination phenotype was verified using independent ZFP3-overexpressing lines, which led to the conclusion that the ZFP3 cDNA encoded C2H2 zinc finger protein and is a novel negative regulator of ABA responses. Enhanced expression of ZFP3 seems to phenocopy the effects of ABA-insensitive abi mutations during germination. The PP2C-type protein phosphatases ABI1 and ABI2 are part of the ABA receptor complex and negatively regulate ABA responses, while the transcription factors ABI3, ABI4, and ABI5 are positive regulators of ABA signals, mediating different aspects of ABA responses (Klingler et al., 2010; Fujita et al., 2011; Finkelstein, 2013). ZFP3 seems to modulate parallel regulatory pathways with ABI2, ABI4, and ABI5, as ZFP3 overexpression in the abi2-1, abi4-101, and abi5-1 backgrounds additively enhances ABA insensitivity conferred by these mutations. ZFP3 may influence ABI5-dependent regulation, as the ABA insensitivity of abi5-1/ZFP3ox lines was only moderately enhanced at high ABA concentrations (Fig. 2).
ZFP3 is a member of a large zinc finger protein family. Although 176 zinc finger proteins have been identified in Arabidopsis (Englbrecht et al., 2004), only a handful of them have been characterized thus far. Similar to ZFP3, overexpression of ZFP1, ZFP4, ZFP6, and ZFP7 led to ABA insensitivity in germination assays and reduced hypocotyl lengths of germinated seedlings (Fig. 2; Supplemental Fig. S14), suggesting their redundant molecular functions. The fact that the double zfp3 zfp4 mutant was more sensitive to ABA than single zfp3 and zfp4 knockouts (Fig. 2) further confirmed that members of this subfamily possess overlapping functions. While ZFP3 expression is rather low in most organs, except roots and vasculature tissues, other members of this gene family can have remarkable activity in particular organs, such as developing seeds (ZFP2), rosette leaves (ZFP8), and inflorescence stems (ZFP6; Supplemental Fig. S7). As is the case for ZFP3, several ZFP genes are down-regulated by ABA, confirming negative interaction between these genes and ABA regulation (Fig. 4; Supplemental Fig. S8). Therefore, alterations in transcriptional regulation can lead to certain differences in biological functions of the individual ZFP genes.
While ZFP3 has not yet been characterized in detail, some of the related factors were reported to control different aspects of plant development. ZFP1 was previously implicated in photomorphogenic responses, although the precise function of this gene was not revealed (Chrispeels et al., 2000). ZFP2 was shown to control floral abscission, as overexpressing plants enhanced the retention of floral organs (Cai and Lashbrook, 2008). ZFP5, ZFP6, and ZFP8 were shown to regulate the differentiation of trichomes through integrating GA and cytokinin signaling (Zhou et al., 2011, 2013). We did not observe abnormalities in the trichome morphology of ZFP3ox or zfp3 mutant seedlings (data not shown). In our experimental system, ZFP6 overexpression resulted in ABA insensitivity and shorter hypocotyls, while ZFP8 had only a minor influence on these traits. Overexpression of ZFP10 and ZFP11 in Arabidopsis and tobacco (Nicotiana tabacum) resulted in dwarf plants with abnormal morphology and reduced fertility (Dinkins et al., 2002, 2012). Similar features were characteristic of ZFP3-overexpressing plants, which were semidwarf with severely compromised fertility (Fig. 3). Although the number of seeds in ZFP3-overexpressing plants was reduced (Supplemental Fig. S5), their size and morphology were similar to those of wild-type plants. Germination of ZFP3-overexpressing seeds in one-half-strength Murashige and Skoog medium was reduced, while it showed ABA insensitivity similar to the A44 line (Fig. 1; Supplemental Fig. S6). These data suggest that ZFP3 interferes with fertilization and seed germination. Ectopic expression of ZFP3-related C2H2 zinc finger proteins resulted in pleiotropic morphological abnormalities, indicating that they are important regulators of plant development. Phenotypes of the zfp3 and zfp4 mutants were similar to that of wild-type plants, suggesting a certain degree of redundancy in the function of related ZFP genes. The fact that constitutive overexpression of ZFP3 impaired fertility justified the strategy to use regulated expression of the cDNA inserts in the COS (Papdi et al., 2008; Rigó et al., 2012), as uncontrolled overexpression of ZFP3 could have prevented the identification and/or recovery of fertile plants during the screen.
Transcript profiling data showed that ZFP3 modulates the expression of a set of ABA- and light-regulated genes. Transcript levels of a number of ABA-induced genes, including late embryogenesis abundant and dehydrin genes, were lower in ZFP3ox seedlings than in the wild type when germinated in the presence of ABA (Table I; Fig. 6; Supplemental Data Set S1). These experiments, however, could not identify unambiguously the primary targets of ZFP3, which will require further studies, including chromatin immunoprecipitation combined with massively parallel sequencing, protein interaction studies, and DNA-binding assays. Reduced expression of ABA-regulated transcription factors such as ABI4 could contribute to the ABA insensitivity of ZFP3ox lines (Fig. 6). The ABA insensitivity and tolerance to paclobutrazol of ZFP3ox lines could be influenced by reduced transcription of the DELLA factor RGL2 in ZFP3ox seedlings, which is a negative regulator of GA responses (Lee et al., 2002; Tyler et al., 2004). RGL2 was shown to control germination by enhancing ABA-dependent ABI3 and ABI5 activities, which act as final repressors of germination (Piskurewicz et al., 2008, 2009). Although the transcript levels of ABI5 and RGL2 were lower in ZFP3ox seedlings, they are probably not direct targets of ZFP3, as their expression was not altered by ZFP3 overexpression after short-term ABA induction (Fig. 6I). Expression of ABI4, however, was reduced by ZFP3ox, suggesting that this AP2/Ethylene Responsive Factor (ERF)-type transcription factor can more directly be connected to developmental consequences of ZFP3 overexpression (Finkelstein et al., 1998; Wind et al., 2013). ABI4 regulates the expression of numerous ABA-induced genes, often in synergistic action with other transcription factors such as ABI5 (Reeves et al., 2011). Therefore, the germination of ZFP3-overexpressing seeds may be promoted by down-regulation of ABI4 and the modulation of expression of a set of genes controlled by this transcription factor (Wind et al., 2013). ABI4 was shown to inhibit the expression of photosynthetic genes (Acevedo-Hernández et al., 2005). Enhanced expression of photosynthetic genes such as CAB2 in ZFP3ox seedlings, therefore, can be the consequence of reduced ABI4 levels (Table II; Fig. 6; Supplemental Data Set S1). Nevertheless, ABI4 is likely only one of the targets of ZFP3 regulation, as ZFP3ox can still enhance ABA insensitivity in the abi4 mutant background.
Seed germination and seedling development are controlled by other factors, such as light (Seo et al., 2009). Light signals are perceived by different receptors, of which phytochromes are dominant photoreceptors of red and far-red light, playing important roles in the regulation of germination and photomorphogenesis, including the inhibition of hypocotyl elongation (Franklin and Quail, 2010; Li et al., 2011). Our data show that ZFP3 and the closely related ZFP-type factors inhibit hypocotyl elongation, displaying short hypocotyls in light and in darkness. However, this is not accompanied by other photomorphogenic traits like the expansion of cotyledons or the disappearance of the hypocotyl hook (Fig. 7A). This suggests that overexpression of ZFP3 per se does not trigger light responses without illumination. Externally added GA3 and epibrassinolide were unable to complement the short-hypocotyl phenotype of ZFP3ox seedlings, suggesting that ZFP3 acts independently of the signaling networks of these growth regulators.
ZFP3 interferes with red light but not far-red or blue light signaling, manifested by the inhibition of hypocotyl elongation specifically under red light. This suggests that ZFP3 functions as an amplifier of light signals originating from PhyB, PhyC, PhyD, or PhyE receptors but does not affect PhyA-mediated signaling. The fact that the long hypocotyls of phyB mutant seedlings were considerably reduced by ZFP3 overexpression in red light (Fig. 8) indicated that ZFP3 can modulate photomorphogenic signals derived from red light receptors other than PhyB. Thus, we propose that ZFP3 is a positive component of the red light signaling pathway(s). PIF transcription factors are negative regulators of red light signaling and promote scotomorphogenesis in dark-grown seedlings, controlling the expression of a large set of target genes (Leivar et al., 2012; Zhang et al., 2013). While transcript levels of PIF genes were not altered in ZFP3ox seedlings, many genes that were repressed or derepressed by ZFP3 overexpression were down- and up-regulated in the quadruple pif1,3,4,5 mutant, respectively (Table III; Supplemental Data Sets S2–S5; Leivar et al., 2012). These data indicate that a subset of ZFP3-regulated genes are also targets of PIF-dependent transcriptional regulation, suggesting that ZFP3 and PIFs control an overlapping signaling pathway.
While a close relationship between light and GA signaling in early seedling development is well documented (Seo et al., 2009; Lau and Deng, 2010), the role of ABA in light-regulated seedling morphogenesis is not well known. The ABI5 transcription factor is a component of the ABA signaling pathway in Arabidopsis, whose expression is controlled by the light signaling component HY5 and can shorten hypocotyls in a light-dependent manner if overexpressed in Arabidopsis (Chen et al., 2008). While ectopic expression of ZFP3 in the Col-0 background produced seedlings with short hypocotyls, light-dependent hypocotyl elongation in the abi5-1 mutant background was only minimally affected (Fig. 8; Supplemental Fig. S17). This demonstrates that ZFP3 exerts its effect on light-controlled hypocotyl elongation exclusively through ABI5. Overexpression of ABI5 shortened hypocotyls under red, far-red, and blue light illumination (Chen et al., 2008), whereas the control of light-dependent hypocotyl elongation by ZFP3 was more specific to red light.
Our study shows that ZFP3, together with closely related ZFP factors, is a negative regulator of ABA signaling during germination and early seedling development (Supplemental Fig. S18). Upon overexpression of ZFP3, seeds can germinate in the presence of inhibitory concentrations of ABA. Moreover, ZFP3 strengthens red light signals perceived by photoreceptors other than PhyA, leading to reduced hypocotyl elongation of germinating seedlings. ABI5 seems to be epistatic to ZFP3 in the control of red light-dependent repression of hypocotyl elongation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) overexpression and mutant lines were derived from ecotype Col-0 unless mentioned otherwise. Arabidopsis growth conditions in sterile culture and controlled growth chambers were as described earlier (Koncz et al., 1994). Plant transformation was performed using the in planta Agrobacterium tumefaciens infiltration method (Bechtold et al., 1993). Arabidopsis cell suspensions were established, maintained, and transformed as described (Ferrando et al., 2000). The COS was used to identify the ABA-insensitive A44 line as described (Papdi et al., 2008).
Phenotyping
Germination assays were performed on one-half-strength Murashige and Skoog medium containing 0.5% (w/v) Suc, solidified with 0.8% (w/v) agar. Sterilized seeds were stratified at 4°C in the dark for 3 d, plated on agar-solidified germination medium, and incubated in a 16-h/8-h light/dark cycle at 22°C. Germination rate was scored by green cotyledon emergence for 15 d. Each plate contained 50 to 100 seeds, and every experiment was repeated three times.
For testing hypocotyl elongation, seeds were sown on wet filter paper stratified for 3 d in darkness at 4°C. Cold-treated seeds were irradiated with white light for 3 h at 22°C and transferred to darkness for an additional 18 h at 22°C. The plates were subsequently placed under various light conditions for 4 d (light source, SNAP-LITE LED lighting system (Quantum Devices Inc.); maximal spectral outputs, blue at 470 nm, red at 670 nm, and far red at 735 nm; Bauer et al., 2004). Hypocotyl elongation was measured on digital images using ImageJ software (rsbweb.nih.gov/ij). Hypocotyl lengths at different fluences of light were normalized to the corresponding dark-grown hypocotyl length to reflect the light-dependent regulation (Bauer et al., 2004). Thirty seedlings were measured in each assay, and the experiments were repeated three times.
Genotyping of zfp Mutants and Genetic Crosses
All the T-DNA mutants were obtained from the Arabidopsis Biological Resource Center and genotyped by PCR using gene-specific primers and left border T-DNA primers (Supplemental Table S1). The zfp3 (At5g25160) GABI_177E02 mutant carried a T-DNA insertion at 25 bp downstream of ATG. The zfp4 (SALK_038923) mutant had an insertion in the 5′ untranslated region at 38 bp upstream of ATG of the At1g66140 gene. The zfp3 mutant was crossed with the zfp4 mutant, and homozygous zfp3 zfp4 double mutants were used for testing. Genetic crosses were made to obtain ZFP3 overexpression in the abi2-1, abi5-1, abi4-101, and phyB-9 backgrounds.
Gene Cloning and Vector Construction
cDNA inserts from transgenic plants were cloned and identified as described (Papdi et al., 2008). Full-length cDNA clones of ZFP transcription factors were obtained from the REGIA collection (Paz-Ares, 2002). Cloned cDNAs were transferred with LR Clonase (Life Technologies) reaction into destination vectors pER8-GW and pB2GW7, which carried inducible and constitutive expression cassettes, respectively.
To construct the ZFP3 promoter-GUS gene fusion, a fragment of the 2.4-kb 5′ upstream region of the AT5G25160 gene was PCR amplified from the BAC clone F21J6 using pZFP3a and pZFP3b primers carrying BamHI and EcoRI cloning sites, respectively (Supplemental Table S1). The PCR fragment was cloned into pBSK(+) vector, the nucleotide sequence was verified, and the error-free promoter fragment was subcloned into pENTR-2B vector and transferred into the pMDC162 promoter testing vector using LR Clonase reaction (Curtis and Grossniklaus, 2003).
To have ZFP3 fused to the HA epitope at the C-terminal and N-terminal ends, cDNA of ZFP3 was cloned in frame into pPILY vector at NcoI and BglII sites for C-terminal tagging and in pMENCHU at SalI and EcoRI sites for N-terminal tagging (Ferrando et al., 2000). The ZFP3-HA (C-terminal) and HA-ZFP3 (N-terminal) gene fusions were subcloned into binary vector pPR97 (Szabados et al., 1995). To create a C-terminal fusion of ZFP3 with GFP, full-length cDNA was PCR amplified with restriction enzyme sites SalI and EcoRI (without stop codons), cloned in pENTR-2B vector, and subcloned into p7FWG2 (Karimi et al., 2002) using LR Clonase reaction. Primers used for cloning and PCR experiments are listed in Supplemental Table S1.
qRT-PCR Analysis
For qRT-PCR analysis, total RNA was isolated from Arabidopsis seedlings with the Qiagen RNeasy Mini Kit. Five micrograms of RNA was treated with Ambion’s TURBO DNA-free DNase, and 1 µg of RNA was used for cDNA synthesis using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR was performed on 5× diluted cDNA templates using the Sigma SYBR Green RT-PCR Reagent Kit and the ABI PRISM 7900 HT Sequence Detection System. GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C2 (At1g13440) was used as an internal reference. Experiments were repeated with three biological replicates.
RNA Sequencing
For RNA sequencing analysis, ZFP3ox and Col-0 wild-type seeds were germinated on medium supplemented with either 5 μm estradiol or 5 μm estradiol and 2.5 μm ABA. Seedlings on estradiol-containing medium were harvested after 36 h, and seedlings germinated in the presence of ABA and estradiol were harvested after 84 h to obtain seedlings in a physiologically identical radicle stage for both conditions. Total RNA was isolated with the Qiagen Plant RNeasy Kit, followed by DNaseI treatment using Ambion’s TURBO DNA-free kit. RNA quality and quantity measurements were performed on Bioanalyzer (Agilent Technologies). RNA samples were processed using the SOLiD total RNA-Seq Kit (Life Technologies), with the templates sequenced on the SOLiD 5500xl Instrument using the 50-base sequencing chemistry. Raw, trimmed, 50-base sequences were mapped in a strand-specific way onto the Arabidopsis reference genome. Data were processed with Baggerley’s and Kal’s test with Bonferroni and false discovery rate correction, and the number of sequencing reads generated from each sample was converted into reads per kilobase of exon model per million mapped reads (Mortazavi et al., 2008). Data were submitted to the Gene Expression Omnibus under accession number GSE48661. Coexpression analysis of ZFP3-regulated genes was made with the hierarchical clustering tool of Genevestigator (www.genevestigator.com/gv; Zimmermann et al., 2004).
Measurement of ABA Content
Seven-day-old seedlings were frozen in liquid nitrogen and stored in −80°C until processing. Thirty-milligram samples were extracted in triplicate. ABA content was determined with liquid chromatography-tandem mass spectrometry as described (López-Carbonell et al., 2009). The HPLC system consisted of a Perkin-Elmer series 200 quaternary pump equipped with a thermostated (4°C) autosampler. For extract analysis, we employed an XBridge C18 column (Waters) equipped with a cartridge system coupled to the HPLC column that protects and extends the lifetime of the column by capturing sample contaminants (Securityguard C18 Phenomenex). Mass spectrometry and tandem mass spectrometry analyses were performed on an API 3000 triple quadrupole mass spectrometer (PE Sciex). All the analyses were performed using the Turbo Ionspray source in negative ion mode.
Cellular Technologies
GFP localization was performed on roots of p7FWG2-ZFP3-expressing seedlings using an Olympus confocal laser scanning microscope as described (Székely et al., 2008). Histochemical detection of pZFP3-GUS activity was performed as reported (Jefferson et al., 1987). Images were taken with Nikon SMZ800 microscope equipped with a Nikon Coolpix995 digital camera.
Protein Extraction and Determination
To prepare total protein extracts, plant material was ground in liquid nitrogen in buffer containing 50 mm Tris-Cl (pH 7.5), 0.5% (w/v) SDS, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride with protease inhibitor cocktail (Sigma). Thirty-microgram protein samples were separated on 10% SDS-polyacrylamide gels (Schägger and von Jagow, 1987), blotted to polyvinylidene fluoride membranes by wet electroblotting (Mini-Protean II system; Bio-Rad), and then incubated with a peroxidase-coupled monoclonal anti-HA antibody (Roche). The HA epitope-tagged proteins were detected using the Lumi-Light western-blotting substrate (Millipore).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic tree and sequence alignment of ZFP3-related proteins.
Supplemental Figure S2. Germination assay of independent ZFP3ox lines.
Supplemental Figure S3. Characterization of zfp3 and zfp4 mutants.
Supplemental Figure S4. Complementation of the zfp3 mutant.
Supplemental Figure S5. Constitutive overexpression of ZFP3 reduces fertility.
Supplemental Figure S6. Constitutive overexpression of ZFP3 reduces seed germination.
Supplemental Figure S7. Developmental regulation of 10 ZFP-related genes.
Supplemental Figure S8. Hormonal regulation of 10 ZFP-related genes.
Supplemental Figure S9. Gene Ontology terms of genes differentially regulated by ABA and ZFP3.
Supplemental Figure S10. Hierarchical clustering of ZFP3-repressed gene sets.
Supplemental Figure S11. Hierarchical clustering of ZFP3-induced gene sets.
Supplemental Figure S12. Hierarchical clustering of ABA-induced and ZFP3-repressed gene sets.
Supplemental Figure S13. Hierarchical clustering of ABA-repressed and ZFP3-derepressed genes.
Supplemental Figure S14. Hypocotyl elongation of Arabidopsis lines overexpressing cDNAs of ZFP3-related genes.
Supplemental Figure S15. Effect of plant hormones on the hypocotyl elongation of wild-type, ZFP1ox, and ZFP3ox plants.
Supplemental Figure S16. Effect of plant hormones on the germination of ZFP3ox and zfp3 zfp4 double mutant seeds.
Supplemental Figure S17. Hypocotyl elongation of abi mutant lines overexpressing ZFP3.
Supplemental Figure S18. Regulatory functions of ZFP3 in seed germination and plant development.
Supplemental Table S1. List of primers and oligonucleotides.
Supplemental Text S1. Description of hierarchical clustering and mutants listed in Table III.
Supplemental Data Set S1. Differentially regulated genes identified in the transcript profiling experiment.
Supplemental Data Set S2. Hierarchical clustering of ZFP3-down-regulated genes.
Supplemental Data Set S3. Hierarchical clustering of ZFP3-up-regulated genes.
Supplemental Data Set S4. Hierarchical clustering of ABA-induced and ZFP3-down-regulated genes.
Supplemental Data Set S5. Hierarchical clustering of ABA-repressed and ZFP3-up-regulated genes.
Supplementary Material
Acknowledgments
We thank Annamári Király and Mihály Dobó for growing the plants, Ferhan Ayaydin for assistance in microscopy, Balázs Horváth for contributing to transcript profiling, and Ferenc Nagy for valuable discussion and correction of the article.
Glossary
- ABA
abscisic acid
- cDNA
complementary DNA
- COS
conditional cDNA-overexpressing system
- T-DNA
transfer DNA
- qRT
quantitative real-time
- Col-0
Columbia-0
- ACC
1-aminocyclopropane-1-carboxylic acid
Footnotes
This work was supported by Országos Tudományos Kutatási Alap (grant no. K–81765), Hungary-Serbia Cross-border Co-operation Programme (project no. HUSRB/1002/214/036), and European Cooperation in Science and Technology Action (action no. FA0605).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Acevedo-Hernández GJ, León P, Herrera-Estrella LR. (2005) Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. Plant J 43: 506–519 [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris 316: 1194–1199 [Google Scholar]
- Cai S, Lashbrook CC. (2008) Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol 146: 1305–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M. (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373–383 [DOI] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L. (2008) Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci USA 105: 4495–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels HE, Oettinger H, Janvier N, Tague BW. (2000) AtZFP1, encoding Arabidopsis thaliana C2H2 zinc-finger protein 1, is expressed downstream of photomorphogenic activation. Plant Mol Biol 42: 279–290 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins R, Pflipsen C, Thompson A, Collins GB. (2002) Ectopic expression of an Arabidopsis single zinc finger gene in tobacco results in dwarf plants. Plant Cell Physiol 43: 743–750 [DOI] [PubMed] [Google Scholar]
- Dinkins RD, Tavva VS, Palli SR, Collins GB. (2012) Mutant and overexpression analysis of a C2H2 single zinc finger gene of Arabidopsis. Plant Mol Biol Rep 30: 99–110 [Google Scholar]
- Englbrecht CC, Schoof H, Böhm S. (2004) Conservation, diversification and expansion of C2H2 zinc finger proteins in the Arabidopsis thaliana genome. BMC Genomics 5: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando A, Farràs R, Jásik J, Schell J, Koncz C. (2000) Intron-tagged epitope: a tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J 22: 553–560 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book 11: e0166, doi/10.1199/tab.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Somerville CR. (1990) Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol 94: 1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fohgrub U, Kempken F. (2012) Molecular analysis of fungal gene expression upon interkingdom competition with insects. Methods Mol Biol 944: 279–286 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Shinozaki K, Yamaguchi-Shinozaki K. (2011) ABA-mediated transcriptional regulation in response to osmotic stress in plants. J Plant Res 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Gan S. (2004) A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis. Plant Mol Biol 54: 1–9 [DOI] [PubMed] [Google Scholar]
- Hsieh WP, Hsieh HL, Wu SH. (2012) Arabidopsis bZIP16 transcription factor integrates light and hormone signaling pathways to regulate early seedling development. Plant Cell 24: 3997–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Klingler JP, Batelli G, Zhu JK. (2010) ABA receptors: the START of a new paradigm in phytohormone signalling. J Exp Bot 61: 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Martini N, Szabados L, Hrouda M, Bachmair A, Schell J (1994) Specialized vectors for gene tagging and expression studies. In SB Gelvin, ed, Plant Molecular Biology Manual, Vol B2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–22 [Google Scholar]
- Koornneef M, Reuling G, Karssen CM. (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377–383 [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. (2006) The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol 140: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X. (2011) Phytochrome signaling mechanisms. The Arabidopsis Book 9: e0148, doi/10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Carbonell M, Gabasa M, Jáuregui O. (2009) Enhanced determination of abscisic acid (ABA) and abscisic acid glucose ester (ABA-GE) in Cistus albidus plants by liquid chromatography-mass spectrometry in tandem mode. Plant Physiol Biochem 47: 256–261 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Merlot S, Mustilli AC, Genty B, North H, Lefebvre V, Sotta B, Vavasseur A, Giraudat J. (2002) Use of infrared thermal imaging to isolate Arabidopsis mutants defective in stomatal regulation. Plant J 30: 601–609 [DOI] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Ohno CK, Reddy GV, Heisler MG, Meyerowitz EM. (2004) The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Papdi C, Abrahám E, Joseph MP, Popescu C, Koncz C, Szabados L. (2008) Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol 147: 528–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Giraudat J. (1997) Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J 11: 693–702 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J. (2002) REGIA, an EU project on functional genomics of transcription factors from Arabidopsis thaliana. Comp Funct Genomics 3: 102–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. (2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Turecková V, Lacombe E, Lopez-Molina L. (2009) Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J 28: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR. (2001) The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13: 1263–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WM, Lynch TJ, Mobin R, Finkelstein RR. (2011) Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol Biol 75: 347–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rigó G, Papdi C, Szabados L. (2012) Transformation using controlled cDNA overexpression system. Methods Mol Biol 913: 277–290 [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. (1995) Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203 [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S. (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69: 463–472 [DOI] [PubMed] [Google Scholar]
- Sirichandra C, Wasilewska A, Vlad F, Valon C, Leung J. (2009) The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J Exp Bot 60: 1439–1463 [DOI] [PubMed] [Google Scholar]
- Szabados L, Charrier B, Kondorosi A, deBruijn FJ, Ratet P. (1995) New plant promoter and enhancer testing vectors. Mol Breed 1: 419–423 [Google Scholar]
- Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, et al. (2008) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53: 11–28 [DOI] [PubMed] [Google Scholar]
- Takeda S, Matsumoto N, Okada K. (2004) RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131: 425–434 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. (2004) Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK. (2005) Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans 33: 375–379 [DOI] [PubMed] [Google Scholar]
- Wind JJ, Peviani A, Snel B, Hanson J, Smeekens SC. (2013) ABI4: versatile activator and repressor. Trends Plant Sci 18: 125–132 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH. (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, An L, Sun L, Zhu S, Xi W, Broun P, Yu H, Gan Y. (2011) ZINC FINGER PROTEIN5 is required for the control of trichome initiation by acting upstream of ZINC FINGER PROTEIN8 in Arabidopsis. Plant Physiol 157: 673–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Sun L, Zhao Y, An L, Yan A, Meng X, Gan Y. (2013) Zinc Finger Protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. New Phytol 198: 699–708 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.