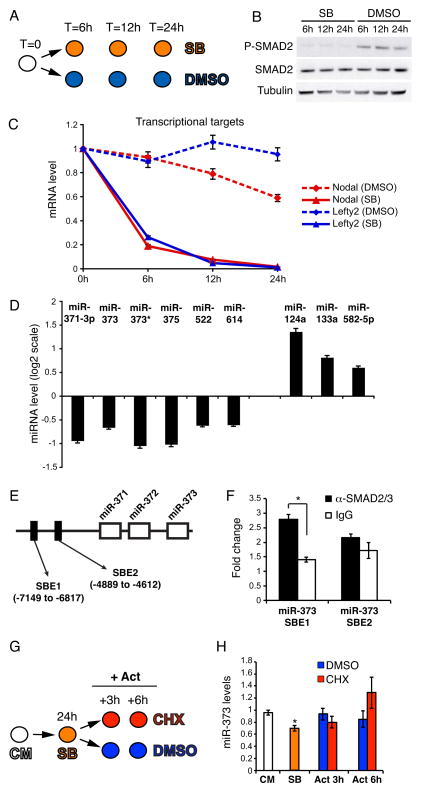
Figure 1. Inhibition of TGFβ signaling in hESCs results in deregulated miRNA expression.
(A) RUES2 cells were cultured under pluripotency conditions in the presence of either 10μM SB431542 (SB) or DMSO, and were harvested at the indicated time points. (B) Western blot showing that SB treatment leads to complete inhibition of C-terminal Smad2/3 phosphorylation (P-SMAD2), without affecting total Smad2/3 protein (SMAD2) levels. β-Tubulin was used as a loading control. (C) Real-Time qPCR analysis of Nodal and Lefty2 reveals a clear inhibition of the pathway. Data points represent average from triplicates. (D) Illumina miRNA microarray analysis of SB-treated RUES2 cells; shown here are the miRNAs whose expression was significantly (p < 0.01; FDR < 0.25) up- or down-regulated after 24 hours of SB treatment. (E) Schematic representation, not in scale, of the miR-371/372/373 locus on chromosome 19. The position of two putative SMAD2/3 binding elements (SBE) [32] is depicted. The numbers are relative to the first nucleotide of pre-miR-371. (F) Real-Time qPCR analysis of the chromatin immunoprecipitated with the indicated antibodies. Fold change is relative to a negative control genomic region on chromosome 4. Data points represent average from triplicates (*p<0.01). (G) RUES2 cells were cultured in the presence of 10μM SB431542 (SB) for 24 hours, then the medium was changed and the cells were cultured for additional 3 or 6 hours in CM containing 50ng/ml ActivinA in absence (DMSO) or presence of 100μg/ml of the protein synthesis cycloheximide (CHX). Cells were harvested at the indicated time points. CM: control untreated cells. (H) Real-Time qPCR analysis of miR-373 expression. Data points represent average from triplicates. * indicates statistically significant difference from the SB sample (p<0.01).