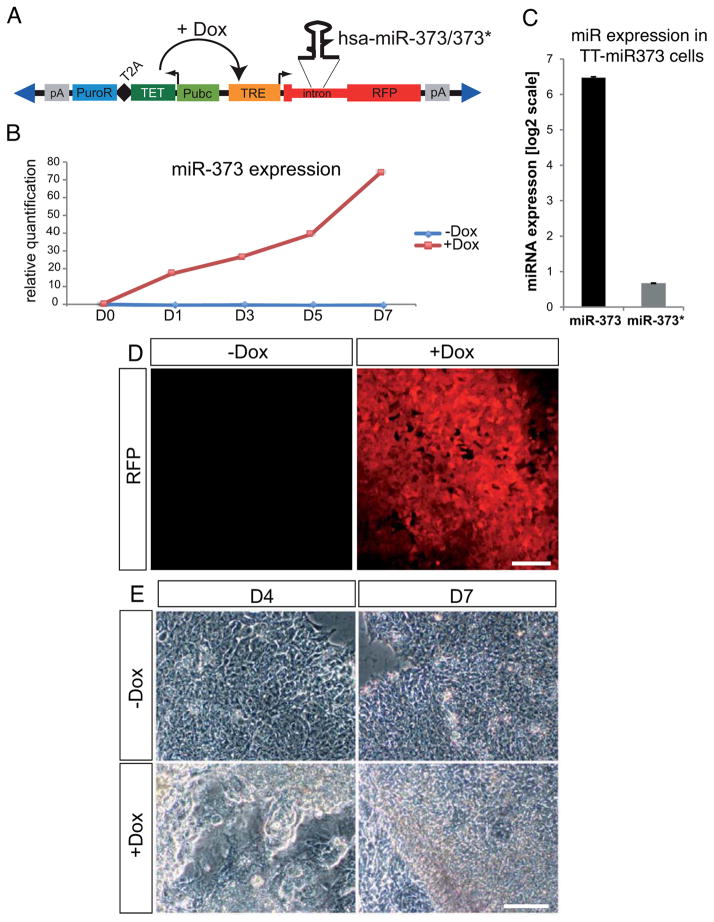
Figure 2. mir-373 overexpression in hESCs using ePiggyBac-mediated transgenesis.
(A) Schematic representation of the ePiggyBac(ePB)-based inducible system used for mir-373 overexpression. RFP: Red Fluorescent Protein gene; pA: polyadenylation signal; PuroR: puromycin resistance gene; T2A: self-cleavage peptide; TET: TET transactivator protein gene; Pubc: human Ubiquitin C constitutive promoter; TRE: TET responsive element; Dox: doxycycline. Blue triangles represent terminal repeats of the transposon. (B) Real-Time miRNA qPCR analysis showing that doxycycline addition induces miR-373 expression compared to control (−dox) cells. Data points are presented as relative quantification to the values at D0. (C) Real-Time miRNA qPCR analysis on TT-mir373 induced (+Dox) cells for five days reveals that miR-373 is the predominantly expressed miRNA from the mir-373 hairpin. Results represent fold-change of expression (at a log2 scale) compared to wild-type RUES2 cells. (D) RFP expression in live TT-mir373 cells five days after addition of either 1μg/ml doxycycline (+Dox) or an equivalent volume of vehicle (H2O) (−Dox). (E) Representative bright-field images from induced (+Dox) or uninduced (−Dox) TT-mir373 cells after four (D4) or seven (D7) days of treatment. Scale bars: 100μm.