Abstract
The skin is a classical example of a tissue maintained by stem cells. However, the identity of the stem cells that maintain the interfollicular epidermis and the source of the signals that control their activity remain unclear. Using lineage tracing and quantitative clonal analyses, we showed that the Wnt target gene Axin2 marks interfollicular epidermal stem cells. These Axin2-expressing cells constitute the majority of the basal epidermal layer, compete neutrally, and require Wnt/β-catenin signaling to proliferate. The same cells contribute robustly to wound healing with no requirement for a quiescent stem cell sub-population. By means of double–labeling RNA in situ hybridization, we showed that the Axin2-expressing cells themselves produce Wnt signals as well as long-range secreted Wnt inhibitors, suggesting an autocrine mechanism of stem cell self renewal.
Stem cells residing in the adult interfollicular epidermis (IFE) regenerate the skin, but the nature of these cells and the molecular signals that regulate them remain incompletely understood. Because of their well-established importance in stem cell maintenance and hair growth, Wnts are candidate self-renewal factors for IFE stem cells. However, Wnt/β-catenin signaling is generally thought to control IFE differentiation rather than self-renewal (1, 2). Reinforcing this view, interfollicular epidermal stem cells (IFESCs) have recently been suggested to originate from more primitive Wnt-independent (Lgr6+) stem cells residing in the hair follicle (3). We sought to dissect the role of Wnt-signaling in IFE homeostasis and regeneration. Because tissue stem cells are commonly influenced by signals secreted by nearby “niche” cells (4), we examined the presence of Wnts and Wnt inhibitors in the skin.
To determine whether Wnt-responding cells are present in the IFE, we looked in mouse skin for cells expressing Axin2, a well-known Wnt/β-catenin target gene. We focused on the mouse hind paw (plantar) epidermis, a region devoid of hair follicles and sweat ducts (Fig. S1A). We marked Axin2-expressing cells using Axin2-CreERT2 and found labeled cells in the basal layer (Figs. 1A, S1E), consistent with Axin2 mRNA and reporter gene expression (Fig. S1B–D). These labeled cells generated clones in multiple IFE compartments that persisted for up to a year (Figs. 1A, S1F), demonstrating that Axin2-CreERT2 labeled keratinocytes are self-renewing stem cells.
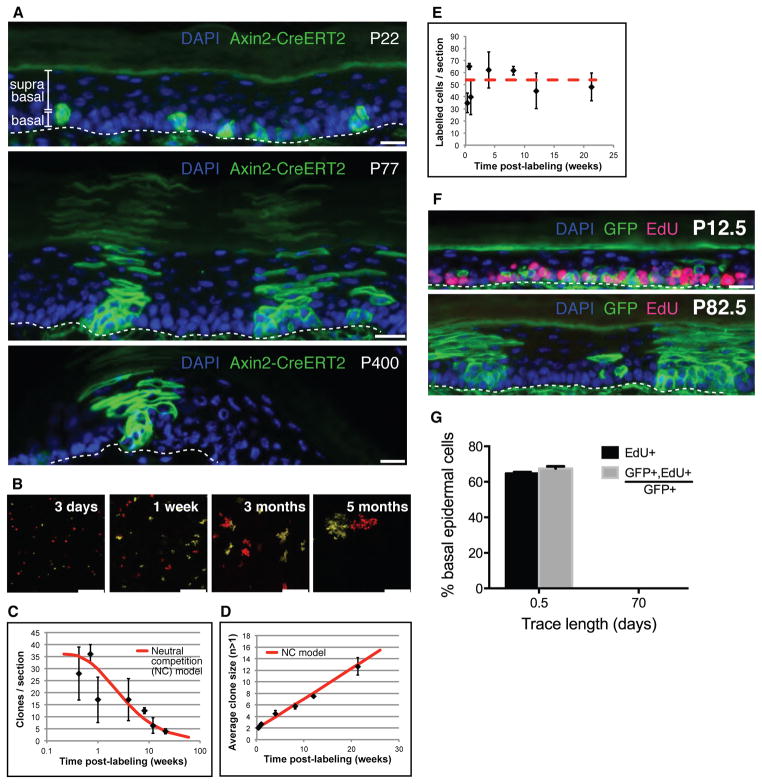
Figure 1. Axin2-expressing basal interfollicular epidermal cells are stem cells that undergo neutral competition and exhibit probabilistic cell fate.
(A) Histological sections of plantar epidermis from Axin2-CreERT2/Rosa26mTmGflox mice chased for 1-day (Postnatal day 22 – P22), 2 months (P77), and more than 1-year (P400). Scale bars, 10 μm. Basal and suprabasal epidermal layers are indicated. Dashed line denotes approximate location of epidermis/dermis boundary. (B) Representative images of whole-mounted Axin2-CreERT2/Rosa26-Rainbowflox plantar epidermis traced from 3 days to 5 months. Only mOrange and mCherry clones in the basal epidermal layer are shown and scored (ST S-II). Scale bars, 100 μm. (C,D) The number of clones per image section drops whereas the average clone size (basal cells per clone) increases, consistent with a model of probabilistic stem cell fate and neutral competition (“NC” model, red curve) (error bars = SD, n>=3 mice). (E) The number of labeled basal cells per image section remains stable; red dashed line shows average over all time points. (F) Representative histological sections of Axin2-CreERT2/Rosa26mTmGflox plantar epidermis chased for 0.5 days (P12.5) and 70.5 days (P82.5) are shown. Dashed line denotes approximate location of epidermis/dermis boundary. Scale bars, 10 μm. (G) Changes in the proportion of EdU+ and GFP+,EdU+/GFP+ basal cells (error bars = SEM). All counts were derived from n>=2 animals per time point and subject to unpaired Student t-tests.
Recent studies examining epidermal stem cell fate provide little indication of the signaling pathways involved in cell fate choice. Using Axin2-CreERT2 as a combined lineage tracing and Wnt reporter tool, we studied the effect of Wnt signaling on cell fate, by analyzing labeled clones at high resolution in whole-mounted epidermis of Axin2-CreERT2/Rosa26-Rainbow (5) mice [Fig. 1B and Supplementary Theory (ST) section S-II]. We first asked whether long-lived Axin2-CreERT2 labeled clones might derive from slow-cycling stem cells that divide with invariant asymmetry to produce transit-amplifying cells (6, 7), or equivalent “committed progenitors” and stem cells that divide with probabilistic fate (8–10). If Axin2-CreERT2 labeled only slow-cycling stem cells dividing with invariant asymmetry, we would expect to see labeled single cells that divide rarely and eventually give rise to stable, long-lived clones. In contrast, the probabilistic differentiation and self-renewal of stem cells and committed progenitors would lead to a rapid drop in the number of clones as a result of neutral clonal competition, with a concomitant increase in the average size of persisting clones to compensate for those that are lost (11). In addition, within a few cell divisions, the size distribution of the persisting clones would follow a simple exponential curve. Comparing the clonal data to these predictions, we found that the labeled Wnt-responding cells and their progeny exhibited all of the characteristics of probabilistic fate and neutral clonal competition (Fig. 1C,D,S2A-C, and ST S-III, S-IV).
To determine whether active Wnt signaling, as indicated by Axin2 expression, occurs in a functionally distinct subpopulation of IFESCs, we examined the number of Axin2-CreERT2 labeled cells in the basal layer over time. Between 3 days and 5 months after initial labeling, the total number of labeled cells in the basal layer of the epidermis remained constant (Pearson correlation coefficient R=0.08 to time after labeling) (Fig. 1E,S2H). This indicates that both Axin2-CreERT2-labeled and unlabeled cells have equal self-renewal capacity in homeostasis, suggesting that all IFESCs express Axin2 (Fig. S1B-D), but only a subset is labeled when treated with Tamoxifen. Further supporting the notion that Axin2-expressing cells are representative of the general population of IFESCs, clonal outcomes showed the same probabilities of division and differentiation at early and late time points (Fig. S2D-E, ST S-V). Thus, Axin2-CreERT2 labeled cells were not biased in their fate choice and were not enriched in a subpopulation of slow-cycling stem cells. If slow-cycling IFESCs are present, they too undergo neutral competition (ST S-VI). However, using a DNA label retaining assay (12, 13) (Fig. S3A), we were unable to detect any label-retaining cells in or outside of persisting Axin2-CreERT2 labeled clones (Fig. 1F,G,S3B-E; ST S-VI).
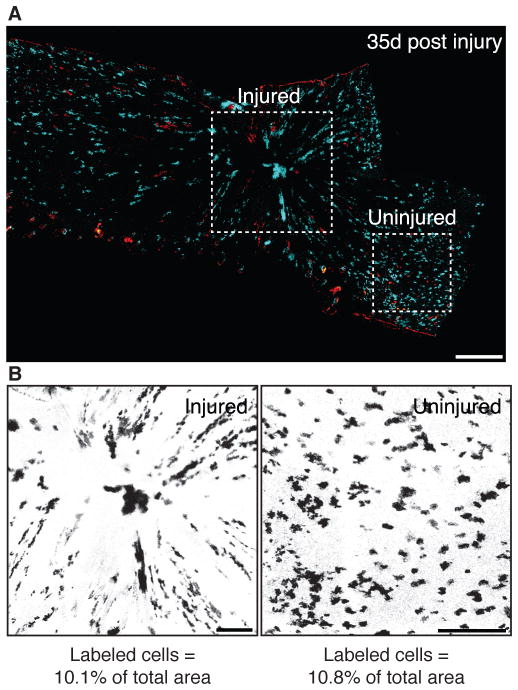
To further test the regenerative potential of Axin2-expressing IFESCs, we induced full-thickness skin biopsy punch wounds in labeled Axin2-CreERT2/Rosa26-mTmGflox mice (Fig. S4A). We found large numbers of relatively even-sized clones radiating into the healed epidermis that persisted for up to 35 days (Fig. 2A,S4B), showing that Axin2-expressing IFESCs robustly contribute to regeneration. However, the labeled cells constituted similar percentages of injured and uninjured skin (Fig. 2B,S4C), indicating that labeled and unlabeled cells have equal abilities to regenerate. Consistent with data from our cell label-retention assays (Fig. 1F,G,S3; ST S-VI), these results also indicate that, if they are present, rare slow-cycling stem cells are not the primary contributors to epidermal wound repair as previously suggested (10).
Figure 2. Axin2-expressing interfollicular epidermal stem cells contribute robustly to wound repair.
(A) Whole-mount views of healing Axin2-CreERT2/Rosa26-Rainbowflox plantar epidermis at 35-days post-wounding. Dashed squares denote approximate injured (left) and uninjured (right) areas. (B) Image masks of injured and uninjured areas. Scale bars, 300 μm.
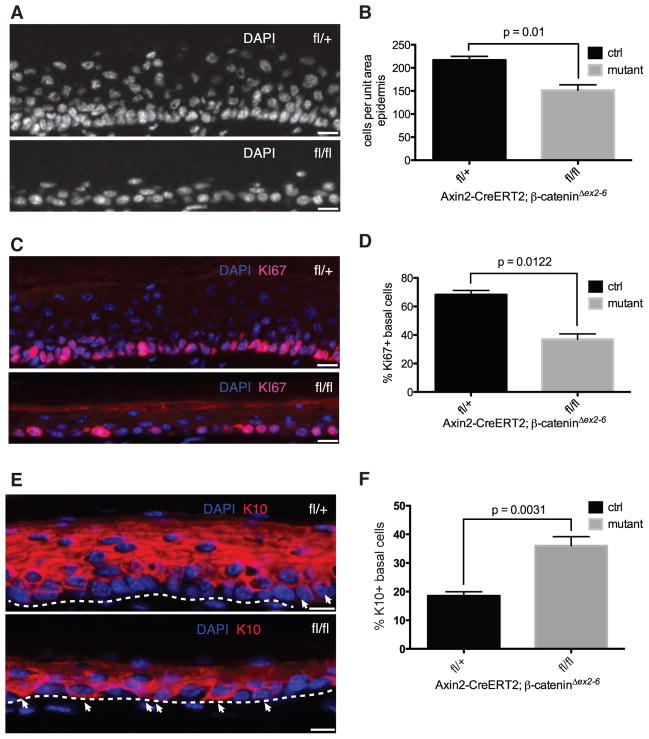
We next tested whether Axin2-expressing IFESCs functionally require Wnt/β-catenin signaling, by conditionally inactivating the gene encoding β-catenin in Axin2-expressing cells. We found an average 30% reduction in the overall cellularity of mutant epidermises (Fig. 3A,B). Consistent with this, 68±3% of control basal cells expressed Ki67 (Fig. 3C,D), a marker of proliferating cells, whereas only 35±4% of mutant basal cells were Ki67-positive (Fig. 3C,D), suggesting a proliferation defect. Similarly, the number of basal cells expressing phosphohistone-H3, another marker of dividing cells, was significantly decreased (Fig. S5A,B). To determine whether epidermal differentiation was also affected, we stained skins for Keratin-10 (K10), an early marker of keratinocyte differentiation. Only 18±1% of control basal cells expressed K10, consistent with estimates obtained from clonal analysis (ST S-IV), whereas 36±1% of mutant basal cells were K10-positive (Fig. 3E,F). Although we cannot exclude systemic effects, our results suggest that IFESCs that are mutant for β-catenin stop proliferating and undergo differentiation. Taken together with our clonal analysis, this suggests that Wnt/β-catenin signaling maintains the IFE stem cell proliferative state but does not affect the likelihood of symmetric self-renewal or differentiation of individual cells.
Figure 3. Axin2-expressing interfollicular epidermal stem cells require β-catenin to proliferate and maintain normal epidermal homeostasis.
(A,C,E) Representative images of DAPI, Ki67 and K10 immunostaining of control Axin2-CreERT2/β-cateninΔex2-6-fl/+ or -del/+ and mutant Axin2-CreERT2/β-cateninΔex2-6-fl/fl or -fl/del plantar epidermis. Arrows in (E) indicate basal epidermal cells staining positive for K10. Dashed lines denote approximate location of epidermis/dermis boundary. (B,D,F) Changes in cellularity, proliferative index, and differentiation between control and mutant plantar epidermises as determined by counting and plotting (B) DAPI+, (D) Ki67+ nuclei and (F) K10+ basal cells (error bars = SEM). All counts were derived from n>=3 independent experiments and subject to pairwise Student t-tests. Scale bars, 10 μm.
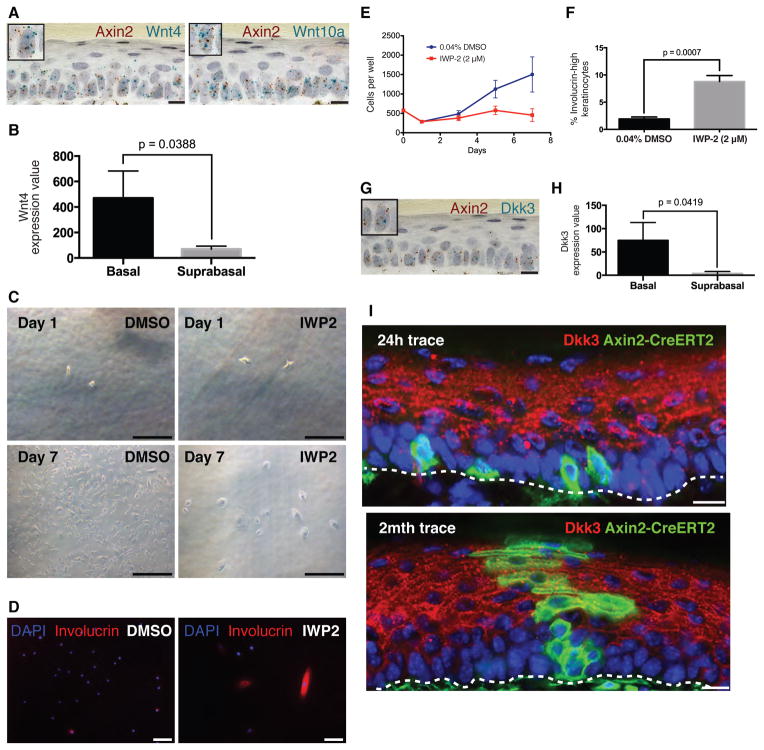
So where do the Wnt signals come from, and how is the niche for IFESCs organized in a way that permits neutral competition? With the use of double–labeling RNA in situ hybridization, we found that Axin2-expressing basal cells in the postnatal epidermis are themselves the source of Wnt signals, expressing several Wnt gene products, including Wnt4 and Wnt10a (Fig. 4A,S6B). This pattern of Wnt gene expression is consistent with previous reports regarding the embryonic basal epidermis (14, 15). Further supporting this observation, primary basal epidermal cells isolated from human skin express Wnt4, whereas suprabasal epidermal cells do not (Fig. 4B) (16). Similarly, cultured primary adult human epidermal keratinocytes express various Wnt genes, as well as Porcupine (Porcn), which is required for Wnt secretion (Fig. S7).
Figure 4. Axin2-expressing interfollicular epidermal stem cells express Wnt and Dkks.
(A) Representative images of double-labeling RNA in situ hybridization in mouse plantar epidermis for Axin2 (red spots) and Wnt4 or Wnt10a (turquoise spots). Inset boxes show magnified view of individual basal cells expressing both Axin2 and Wnts. Scale bars, 10 μm. (B) Wnt4 expression in β4-integrin+ primary human basal epidermal keratinocytes vs β4-integrin- suprabasal epidermal keratinocytes (error bars = SD). Expression values are from GEO dataset GSE26059. (C–D) Representative (C) bright field or (D) immunofluorescence images of keratinocytes continuously cultured in defined medium with either 0.04% DMSO or 2uM IWP2, at the beginning (Day 1) and the end (Day 7) of the experiment, then stained for Involucrin. Scale bars, 50 μm (bright field) or 100 μm (immunofluorescence). (E,F) Changes in the (E) number of cells and (F) percentage of involucrin-high cells per well of keratinocytes treated with either 0.04% DMSO or 2uM IWP-2 (error bars = SEM). Cell counts at all time points were derived from n=3 replicate wells. (G) Representative image of double-labeling RNA in situ hybridization for Axin2 (red spots) and Dkk3 (turquoise spots). Inset box shows magnified view of individual basal cells expressing both Axin2 and Dkk3. Scale bar, 10 μm. (H) Dkk3 expression in primary human β4-integrin+ basal epidermal keratinocytes vs β4-integrin- suprabasal epidermal keratinocytes (error bars = SD). Expression values are from GEO dataset GSE26059. (I) Representative images of Dkk3 immunostaining in plantar epidermises of Axin2-CreERT2/Rosa26mTmGflox mice exposed to Tam at P21 and chased for 1-day (P22) and 2 months (P77). Scale bars, 10 μm.
To determine whether IFESCs functionally require the Wnt that they produce, we treated human epidermal keratinocytes with IWP-2, a validated small-molecule inhibitor of Wnt secretion, and cultured them at clonal density in a defined medium. IWP-2 treated keratinocytes were sparsely distributed and became large and flattened with arrested growth, unlike the densely packed, cuboidally shaped control keratinocytes (Fig. 4C,E). Many more IWP-2-treated keratinocytes also expressed high levels of involucrin, a marker of advanced keratinocyte differentiation (Fig. 4D,F). These data are consistent with our in vivo observations that IFESCs undergo premature differentiation upon loss-of-function mutations in Wnt signaling (Fig. 3E,F).
If IFESCs are both the source and the target of Wnt signals, how might they escape from this autocrine loop and enter a differentiation process? Several genes for secreted Wnt inhibitors, including Dickkopf-1 (Dkk1), Dkk3, and Wnt Inhibitory Factor-1 (WIF1) are expressed in the skin (17–19). With double–labeling RNA in situ hybridization, we saw overlapping expression of Dkks and Axin2 expression in basal cells (Fig. 4G,S6C). This is similar to the situation in human skin, in which primary human basal cells either isolated from skin tissue or cultured in vitro express Dkks (Fig. 4H,S7). Although the Dkk (Fig 4G,H,S6C) and WIF1 (19) mRNAs are mostly located in basal cells, the secreted WIF1 and Dkk3 proteins accumulate at high levels in the suprabasal layers (18, 19). By antibody staining for the Dkk3 protein, we confirmed that Dkk3 is localized to the suprabasal layers, directly adjacent to the Axin2-expressing basal progenitors (Fig. 4I,S8A-H,S9) (18). We tested whether Dkk influences stem cells in the skin by adenoviral overexpression of Dkk, finding that this caused a thinned and hypoproliferative epidermis (Fig. S10) resembling β-catenin mutant skin (Fig. 3A). These data suggest that the differential diffusion of Wnts and Dkk from the basal epidermal stem cells may restrict autocrine Wnt/β-catenin signaling to the basal layer of the epidermis (Fig. S8I). IFESCs leaving the basal layer would encounter increased Wnt-inhibitors and differentiate.
Functional redundancy between the various Wnt inhibitors and Wnts expressed in the skin (Fig.4A,G,S6B,C) may explain the absence of overt phenotypes in mice mutant for these genes (20). However, there is genetic evidence supporting an essential role for Wnt signals in the epidermis. Porcn-knockout mice display a thinned epidermis, similar to that seen in human patients bearing Porcn mutations who develop focal dermal hypoplasia (21–23). Mutations in both Wnt effectors Tcf3 and Tcf4 results in a thinner epidermis (24) whereas deleting β-catenin using the basal epidermal specific driver Keratin-5-rtTA/tet-O-Cre also results in a thinner and hypoproliferative plantar epidermis (25).
Signals emerging from a distinct niche cell compartment are thought to be the main drivers of stem cell self-renewal. We find that epidermal stem cells themselves can be the source of their own self-renewing signals and differentiating signals for their progeny. We postulate that the multiplicity of Wnts and Wnt inhibitors produced by epidermal stem cells allows for fine-tuning of epidermal thickness and wound repair.
Supplementary Material
Acknowledgments
These studies were supported by the HHMI, California Institute of Regenerative Medicine grant TR1-01249, and NIH grants NIH 1U01DK085527, 1R01DK085720, and 5K08DK096048. We thank L. De Simone, A. E. Marcy, and P. H. Chia for cell quantification assistance; C. Logan, S. J. Habib, and A. Oro for manuscript comments; and J. Akech and L.-C. Wang at Advanced Cell Diagnostics for assistance with RNA in situ hybridization. X.L., S.H.T., W.L.C.K., and R.M.W.C. are supported by National Science Scholarships from A*STAR, Singapore. A.M.K. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. K.S.Y. has a Burroughs Wellcome Fund Career Award for Medical Scientists. R.v.A. was supported by a European Molecular Biology Organization long-term fellowship (ALTF 122-2007) and a Dutch Cancer Society fellowship.
References and Notes
- 1.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/S0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 2.Beronja S, Janki P, Heller E, Lien WH, Keyes BE, Oshimori N, Fuchs E. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, Stange DE, Toftgård R, Clevers H. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 4.Losick VP, Morris LX, Fox DT, Spradling A. Drosophila stem cell niches: A decade of discovery suggests a unified view of stem cell regulation. Dev Cell. 2011;21:159–171. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueno H, Weissman IL. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev Cell. 2006;11:519–533. doi: 10.1016/j.devcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Mackenzie IC. Relationship between mitosis and the ordered structure of the stratum corneum in mouse epidermis. Nature. 1970;226:653–655. doi: 10.1038/226653a0. [DOI] [PubMed] [Google Scholar]
- 7.Potten CS. The epidermal proliferative unit: The possible role of the central basal cell. Cell Tissue Kinet. 1974;7:77–88. doi: 10.1111/j.1365-2184.1974.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 8.Clayton E, Doupé DP, Klein AM, Winton DJ, Simons BD, Jones PH. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 9.Doupé DP, Klein AM, Simons BD, Jones PH. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev Cell. 2010;18:317–323. doi: 10.1016/j.devcel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489:257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 11.Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138:3103–3111. doi: 10.1242/dev.060103. [DOI] [PubMed] [Google Scholar]
- 12.Bickenbach JR, McCutecheon J, Mackenzie IC. Rate of loss of tritiated thymidine label in basal cells in mouse epithelial tissues. Cell Prolif. 1986;19:325–333. doi: 10.1111/j.1365-2184.1986.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 13.Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM. Manipulation of stem cell proliferation and lineage commitment: Visualisation of label-retaining cells in wholemounts of mouse epidermis. Development. 2003;130:5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 14.Reddy S, Andl T, Bagasra A, Lu MM, Epstein DJ, Morrisey EE, Millar SE. Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech Dev. 2001;107:69–82. doi: 10.1016/S0925-4773(01)00452-X. [DOI] [PubMed] [Google Scholar]
- 15.Witte F, Dokas J, Neuendorf F, Mundlos S, Stricker S. Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. Gene Expr Patterns. 2009;9:215–223. doi: 10.1016/j.gep.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Radoja N, Gazel A, Banno T, Yano S, Blumenberg M. Transcriptional profiling of epidermal differentiation. Physiol Genomics. 2006;27:65–78. doi: 10.1152/physiolgenomics.00031.2006. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi Y, Itami S, Watabe H, Yasumoto K, Abdel-Malek ZA, Kubo T, Rouzaud F, Tanemura A, Yoshikawa K, Hearing VJ. Mesenchymal-epithelial interactions in the skin: iIncreased expression of dickkopf1 by palmoplantar fibroblasts inhibits melanocyte growth and differentiation. J Cell Biol. 2004;165:275–285. doi: 10.1083/jcb.200311122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du G, Kataoka K, Sakaguchi M, Abarzua F, Than SS, Sonegawa H, Makino T, Shimizu T, Huh NH. Expression of REIC/Dkk-3 in normal and hyperproliferative epidermis. Exp Dermatol. 2011;20:273–277. doi: 10.1111/j.1600-0625.2010.01244.x. [DOI] [PubMed] [Google Scholar]
- 19.Schlüter H, Stark HJ, Sinha D, Boukamp P, Kaur P. WIF1 is expressed by stem cells of the human interfollicular epidermis and acts to suppress keratinocyte proliferation. J Invest Dermatol. 2013;133:1669–1673. doi: 10.1038/jid.2013.42. [DOI] [PubMed] [Google Scholar]
- 20.del Barco Barrantes I, Montero-Pedrazuela A, Guadano-Ferraz A, Obregon M-J, Martinez de Mena R, Gailus-Durner V, Fuchs H, Franz TJ, Kalaydjiev S, Klempt M, Holter S, Rathkolb B, Reinhard C, Morreale de Escobar G, Bernal J, Busch DH, Wurst W, Wolf E, Schulz H, Shtrom S, Greiner E, Hrabe de Angelis M, Westphal H, Niehrs C. Generation and characterization of dickkopf3 mutant mice. Mol Cell Biol. 2006;26:2317–2326. doi: 10.1128/MCB.26.6.2317-2326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrott JJ, Cash GM, Smith AP, Barrow JR, Murtaugh LC. Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proc Natl Acad Sci USA. 2011;108:12752–12757. doi: 10.1073/pnas.1006437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Shaver TM, Balasa A, Ljungberg MC, Wang X, Wen S, Nguyen H, Van den Veyver IB. Deletion of Porcn in mice leads to multiple developmental defects and models human focal dermal hypoplasia (Goltz syndrome) PLoS ONE. 2012;7:e32331. doi: 10.1371/journal.pone.0032331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolognia JL, Jorizzo JL, Schaffer JV. Dermatology. Mosby-Saunders; London: 2012. pp. 869–885. [Google Scholar]
- 24.Nguyen H, Merrill BJ, Polak L, Nikolova M, Rendl M, Shaver TM, Pasolli HA, Fuchs E. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat Genet. 2009;41:1068–1075. doi: 10.1038/ng.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi YS, et al. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.10.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Amerongen R, Bowman AN, Nusse R. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 27.Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22:1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Methods Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 29.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 30.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 31.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preibisch S, Saalfeld S, Tomancak P. Bioinformatics. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein AM, Doupé DP, Jones PH, Simons BD. Kinetics of cell division in epidermal maintenance. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:021910. doi: 10.1103/PhysRevE.76.021910. [DOI] [PubMed] [Google Scholar]
- 37.Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 38.Taylor LR. Aggregation, variance and the mean. Nature. 1961;189:732–735. doi: 10.1038/189732a0. [DOI] [Google Scholar]
- 39.Antal T, Krapivsky PL. J Stat Mech. 2010;7(028):9. [Google Scholar]
- 40.Klein AM, Brash DE, Jones PH, Simons BD. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc Natl Acad Sci USA. 2010;107:270–275. doi: 10.1073/pnas.0909738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.