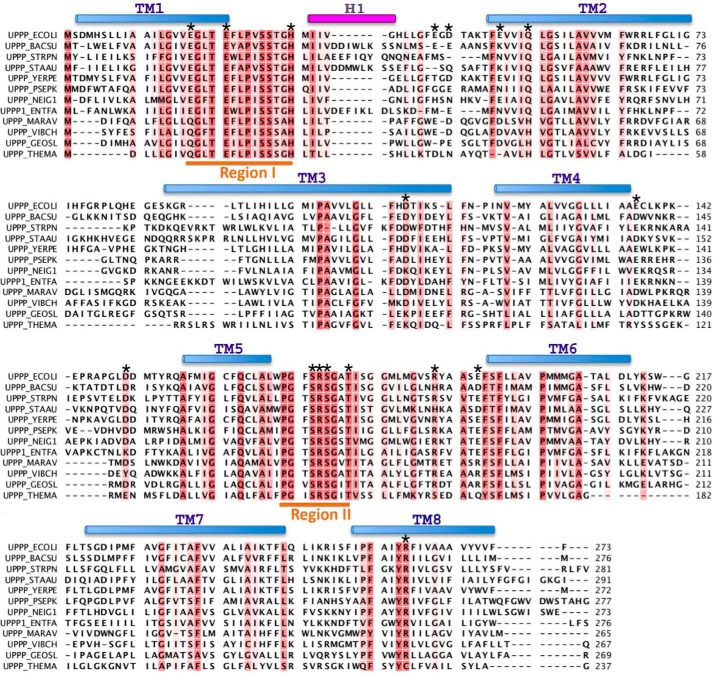
FIGURE 2.
Alignment of the amino acid sequences of the UppP enzymes. The E. coli UppP (UPPP_ECOLI; P60932) is aligned with the following: Bacillus subtilis (UPPP_BACSU; P94507); Streptococcus pneumonia (UPPP_STRPN; P60934); Staphylococcus aureus (UPPP_STAAU; Q9KIN5); Yersinia pestis (UPPP_YERPE; Q8ZI65); Pseudomonas putida (UPPP_PSEPK; Q88IY7); Neisseria gonorrhoeae (UPPP_NEIG1; Q5F6K4); Enterococcus faecalis (UPPP1_ENTFA; Q831R1); Marinobacter aquaeolei (UPPP_MARAV; A1U6U1); Vibrio cholerae (UPPP_VIBCH; Q9KUJ4); Geobacter sulfurreducens (UPPP_GEOSL; P60938); Sulfolobus solfataricus (UPPP_SULSO; Q97X94); and Thermotoga maritima (UPPP_THEMA; Q9WZZ5). The conserved (more than 50%) residues are colored in red. The upper cylinders show the helical regions in the model. The transmembrane (TM) helices are shown in light blue and extracellular helix (H1) is in purple. The two consensus regions I and II are underlined in orange. The residues mutated in this study are indicated by asterisks above the corresponding UppP amino acids.