FIGURE 10.
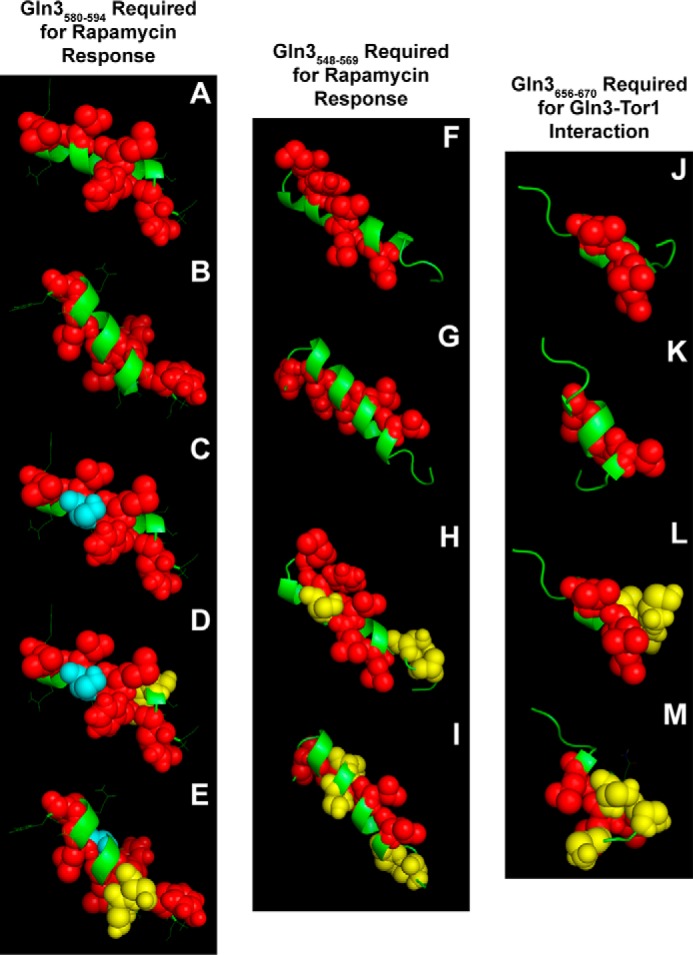
Best estimate structure models of Gln3 sequences with predicted propensities of folding into α-helices. Images A–E (Gln3(580–594)) and F–I (Gln3(548–569)) are required for a response to rapamycin, and images J–M (Gln3(656–670)) are required for a Gln3-Tor1 interaction (40). Red spheres and green ribbons indicate hydrophobic and polar amino acids, respectively. Blue spheres in images C–E indicate the serine residue that was substituted with aspartate or alanine in pRR1194 and pRR1196, respectively. Yellow spheres in images D and E indicate the serine residues that were substituted with aspartate or alanine in pRR1190 and pRR1192, respectively. Yellow spheres in images H and I indicate the serine residues that were substituted with aspartate or alanine in pRR926 and pRR992, and those in images L and M were the residues substituted in pRR850 and pRR1038, respectively (40). Images A, C, D, F, H, J, and L depict one side of the putative helices, and images B, E, G, I, K, and M depict the other side. The models were generated with PEP-FOLD and PHYRE2 and visualized with PyMOL.
