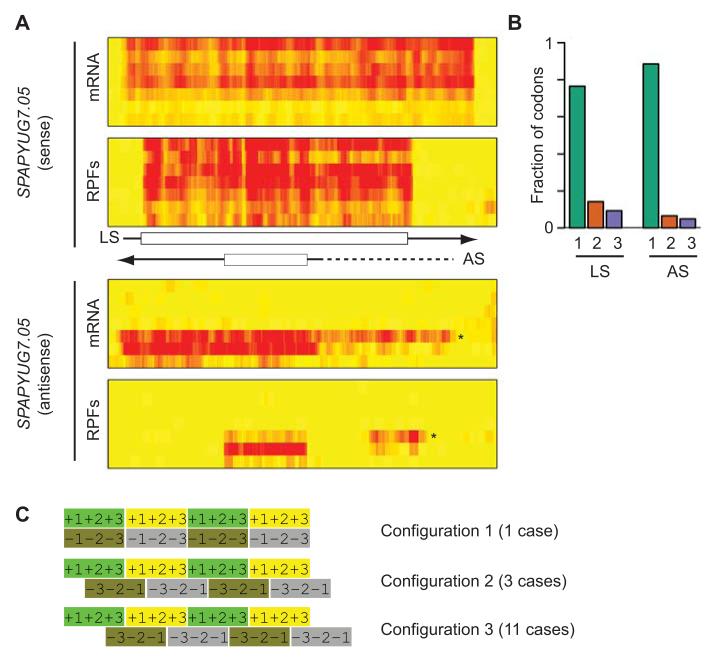
Figure 3. Identification of translated nested antisense genes.
(A) Nested antisense gene in SPAPYUG7.05. The heat maps show mRNA (top) and RPF levels (bottom) across the meiotic time course, starting with vegetative cells and progressing downwards. The brightness of the red color is proportional to the number of reads. The scheme shows the position of UTRs (arrows) and ORFs (boxes). Data for the sense and antisense strands are displayed separately. LS corresponds to the annotated ORF and AS to the newly discovered antisense transcript. The arrows indicate the direction of transcription. Note the presence of a 5′ extension (star on the heat map and dotted line on the scheme) that appears at the 5 hour time point and whose translation correlates inversely with that of the downstream ORF. (B) Triplet periodicity for the sense and antisense ORFs of SPAPYUG7.05. The graphs show the fraction of codons in which the majority of reads map to nucleotides 1, 2 or 3 for the S and L ORFs. (C) Possible relative configurations of reading frames in exonic nested antisense genes. For each configuration the top strand represents translation in the 5′ to 3′ direction, and the bottom one 3′ to 5′. The numbers represent the position of nucleotide 1, 2 and 3 for each codon.