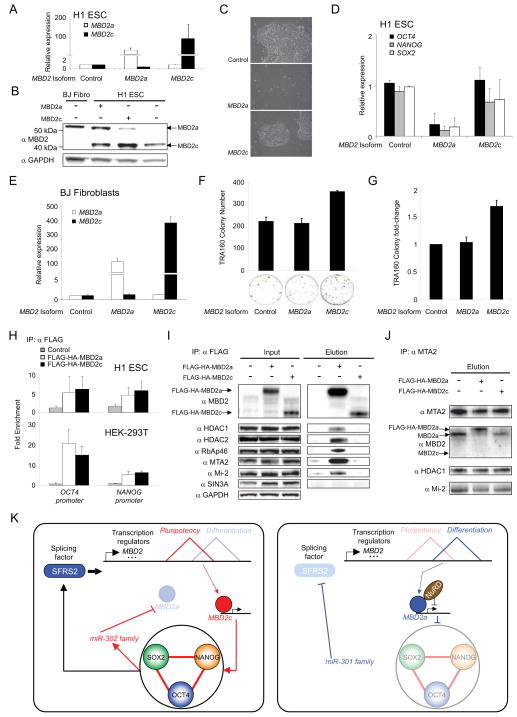
Fig. 4. A general model for regulation of proteome diversity that supports self-renewal in hPSC.
(A–D) Lentiviral-mediated expression of MBD2a but not MBD2c disrupts pluripotency in H1 ESC. Expression of MBD2 isoforms as monitored by (A) qRT-PCR and (B) western blot. Pluripotency in H1 ESC was assessed by (C) colony morphology and (D) expression of OCT4, NANOG, and SOX2. (E–G) Exogenous expression of MBD2c but not MBD2a enhances reprogramming efficiency in BJ DF. (E) Expression of MBD2 isoforms in infected BJ DF as monitored by qRT-PCR. Reprogramming efficiency was assessed by the (F) number and (G) fold change of TRA-1-60 colonies measured across biological replicates. (H) Exogenous MBD2a and MBD2c independently bind to OCT4 and NANOG promoter regions in (top) H1 ESC and (bottom) 293T cells. (I–J) MBD2 interacts with the NuRD complex in an isoform-specific manner. (I) Members of the NuRD transcription repressor complex (HDAC1, HDAC2, MTA2, Mi-2, and RbAp46) co-immunoprecipitate with exogenous FLAG-HA-MBD2a but not FLAG-HA-MBD2c in 293T cells. Neither MBD2 isoform interacts with the SIN3A-Histone deacetylase complex. (J) Co-immunoprecipitation of MTA2 in 293T cells overexpressing FLAG-HA-tagged MBD2 isoforms confirmed the preferential interaction between MBD2a and MTA2, a core NuRD complex member. (K) Proposed model illustrating a putative positive feedback loop, in which the splicing factor SFRS2 along with microRNAs, controlled by the core pluripotency genes, regulate the expression of MBD2 isoforms that either support (MBD2c) or oppose (MBD2a) expression of OCT4, NANOG, and SOX2 through recruitment of the NuRD complex. Error bars represent the mean ± SEM.