Figure 4.
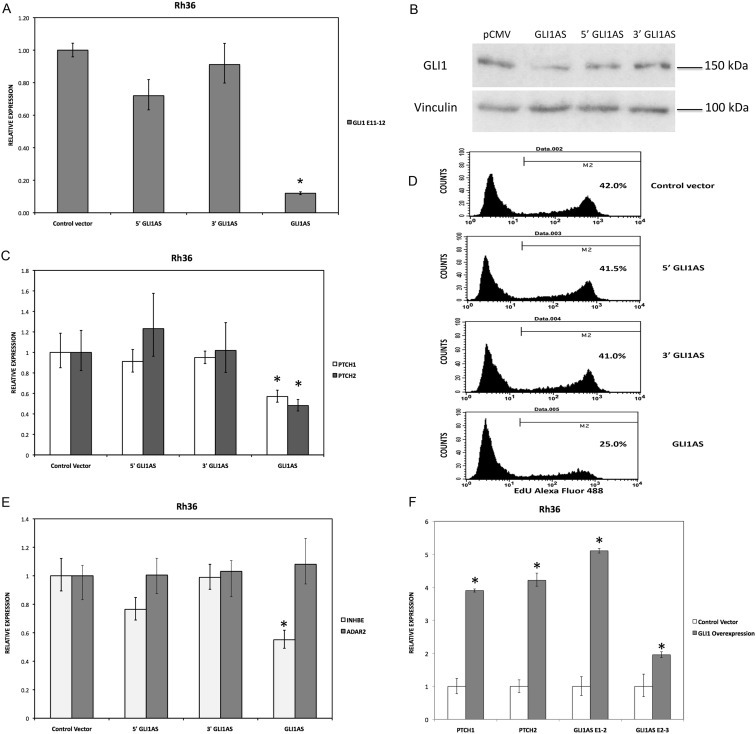
GLI1AS decreases GLI1 expression, while GLI1 increased GLI1AS expression. A. Real‐time RT/PCR analysis of the expression of GLI1 in Rh36 cells, 48 h after transfection of pCMV5 expression constructs for GLI1AS, a 579 bases 5′ segment of GLI1AS (5′GLI1AS) or a 407 bases 3′ segment of GLI1AS (3′GLI1AS). Data are represented as relative expression (2−ΔΔCt values), calculated by subtracting the Ct value of the housekeeping gene TBP from the Ct value of the GLI1 mRNA (ΔCt), and normalized to the ΔCt value obtained following transfection of the pCMV5 control vector, by subtracting this control ΔCt value from the ΔCt values of all samples (ΔΔCt). Error bars indicate the standard deviation. *, Statistical significant, P < 0.01 compared to control, calculated by the Student's t‐test. Note that overexpression of GLI1AS effectively reduced GLI1 expression, while this is not the case when either the 5′ half or the 3′ half of GLI1AS are overexpressed. B. Western blot analysis of GLI1 protein expression in Rh36 cells, following transfection of pCMV5 expression constructs for GLI1AS, 5′GLI1AS or 3′GLI1AS. C. The expression of the typical GLI1 target genes PTCH1 and PTCH2 in Rh36 cells, following transfection of pCMV5 expression constructs for GLI1AS, 5′GLI1AS or 3′GLI1AS, is analyzed as in (A). *, Statistical significant, P < 0.05 compared to control, calculated by the Student's t‐test. D. Rh36 cells 48 h after transfection with pCMV5 control vector, or expression constructs for GLI1AS, 5′ GLI1AS or 3′ GLI1AS were subjected to the EdU incorporation assay for 4 h. The percentage of cells labeled with Alexa Fluor 488 azide and detected by flow cytometry is shown. Note that overexpression of GLI1AS confers an effective reduction of proliferation, while this is not the case when either the 5′ half or the 3′ half of GLI1AS are overexpressed. E. The expression of INHBE, a gene from the GLI1AS locus, and of the unrelated gene ADAR2 in Rh36 cells, following transfection of pCMV5 expression constructs for GLI1AS, 5′GLI1AS or 3′GLI1AS, is analyzed as in (A). *, Statistical significant, P < 0.01 compared to control, calculated by the Student's t‐test. F. The RNA levels of GLI1AS (AS E1‐2 and AS E2‐3), PTCH1 and PTCH2 in Rh36 cells following transfection of a pCMV5 expression construct for GLI1 (Materials and methods) is analyzed as in (A). *, Statistical significant, P < 0.01 compared to control, calculated by the Student's t‐test.
