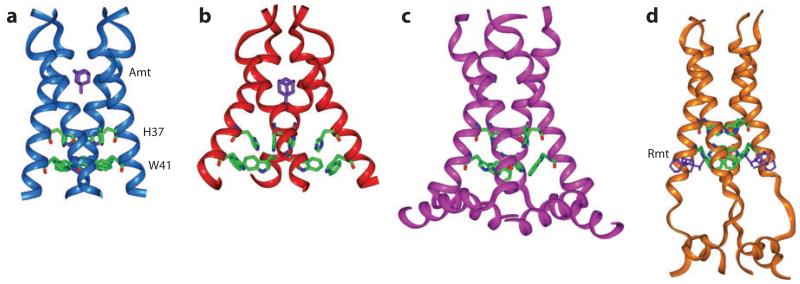
Figure 1.
Various NMR and X-ray crystal structures of the influenza M2 protein. (a) Solid-state NMR structure of M2 (22-46) in DMPC bilayers at pH 7.5 (PDB code: 2KQT). A single amantadine (purple) drug binds to the N-terminal pore. (b) 3.5-Å crystal structure of M2 (25-46) in octylglucoside at pH 5.3 (PDB code: 3C9J). (c) Solid-state NMR orientational structure of M2 (22-62) in DOPC/DOPE bilayers at pH 7.5 (PDB code: 2L0J). (d) Solution NMR structure of M2 (18-60) in DHPC (diheptanoylphosphocholine) micelles at pH 7.5 (PDB code: 2RLF). The four rimantadine drugs (purple) bind to the C-terminal lipid-facing surface of the protein.