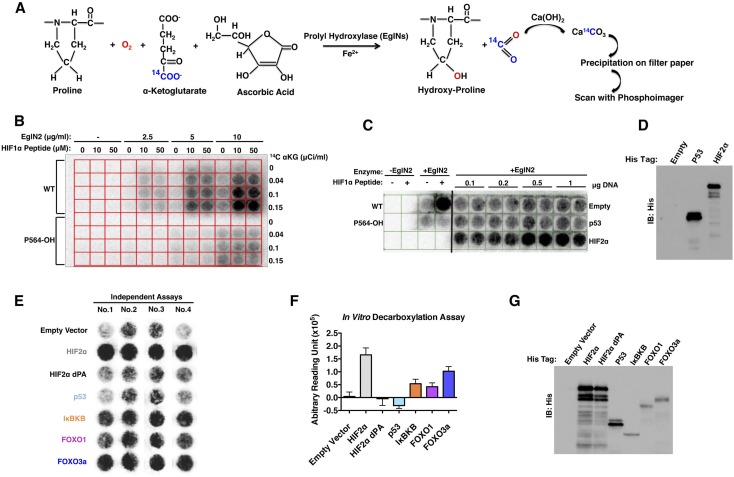
Figure 1.
Screen for EglN2 substrates. (A) Biochemical basis for screen. Hydroxylation of a substrate by EglN2 is coupled to release and capture of 14C-labeled CO2. (B) Hydroxylation assays performed with varying amounts of recombinant EglN2, 14C-αKG, and synthetic HIF1α (556–575) peptides (wild-type or containing hydroxyproline at 564 position). (C) Hydroxylation assays using unprogrammed wheat germ lysate (Empty) or wheat germ lysate programmed to produce His-tagged p53 or HIF2α. Hydroxylation assays with no EglN2 (first two columns) or with EglN2 in the absence or presence of synthetic HIF1α peptides (columns 3,4) were included as controls. Increasing amounts of plasmid DNA were used for in vitro transcription/translation, as indicated. (D) Immunoblot analysis of in vitro translation products used in C. (E) Representative images from four independent hydroxylation assays. (F) Quantification of signal intensity in E (arbitrary units) after subtracting signal intensity from the empty vector well. (G) Immunoblot analysis of in vitro translation products as in E.