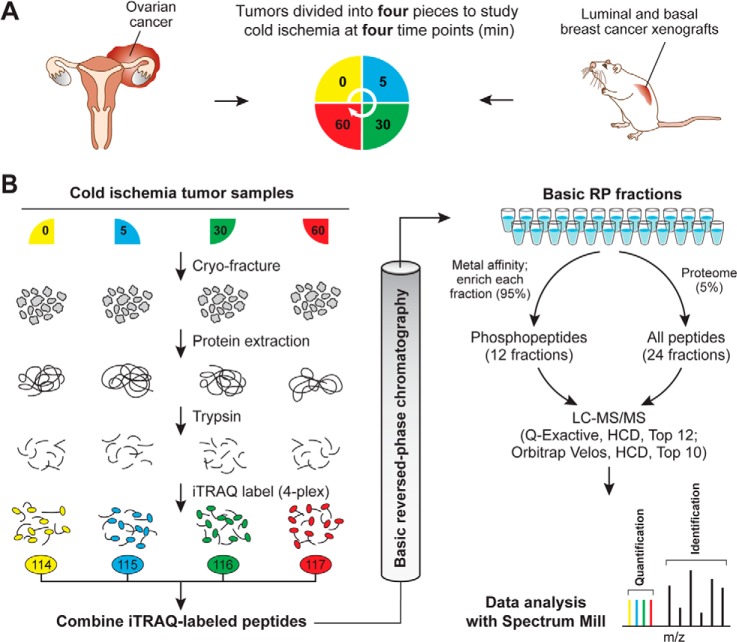
Fig. 1.
Quantitative proteome and phosphoproteome analysis of human ovarian tumors and xenograft breast tumors subjected to controlled ischemia. A, experimental design to study effects of post-excision delay time before freezing across four time points. After excision, tumor samples were cut into four equal pieces and incubated for the indicated times at room temperature before freezing. A total of four different ovarian tumors and three pooled breast cancer xenograft samples for the basal and for the luminal subtype were analyzed. All of these samples were biologically distinct and can be considered as biological replicates. B, quantitative proteomics and phosphoproteomics workflow using 4-plex iTRAQ labeling. Tumor samples were cryofractured and proteins were extracted with urea lysis buffer prior to digestion into peptides using trypsin. Peptide samples derived at four different ischemic time points were labeled using iTRAQ reagents, mixed equably, and separated using high-pH reversed-phase chromatography. Fractions were combined in a noncontiguous way into 24 fractions for proteome analysis (5% of the total material) and 12 fractions for phosphoproteome analysis (95% of the total material). Ovarian cancer samples were analyzed on an LTQ-Orbitrap Velos, and xenograft breast cancer samples were analyzed on a Q Exactive mass spectrometer. Phosphosite and protein identification and quantification were achieved using Spectrum Mill.