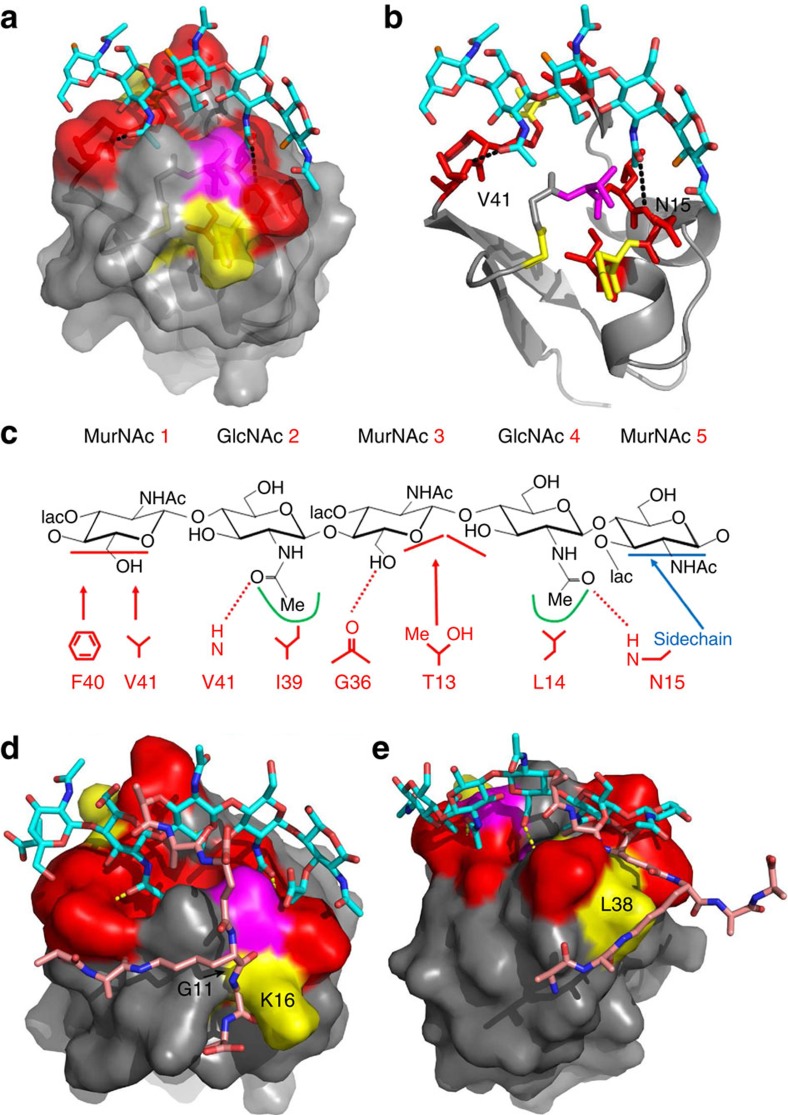
Figure 4. Model of the structure of 1LysM bound to GlcNAc5.
(a) Partially transparent protein surface indicated, showing the groove along the surface, with deeper pockets to accommodate the N-acetyl groups. The O3′ atoms where the MurNAc peptide stems would be attached in a peptidoglycan ligand are shown in orange. (b) Identical view, without the protein surface. Sidechain atoms are shown for T13 and F40. The colour schemes in a and b are the same as in Fig. 3d. (c) Key interactions with MurNAc-(GlcNAc-MurNAc)2. Hydrogen bonds are shown by dashed lines. The horizontal bars indicate that the hydrophobic interaction is to the face of a sugar ring. The two pockets are indicated by green lines. Also shown in blue is the binding location involving N15 discussed in the text which may be important for distinguishing between chitin and peptidoglycan. As discussed in the text, residues I17 and A18 (which show large changes in 15N HSQC; Fig. 3) are buried and do not interact directly with the ligand, but move because of conformational rearrangement. The shift change seen for D37 HN is due to the interaction with G36 carbonyl. (d,e) Model of the interaction between 1LysM and peptidoglycan with peptide stem. The peptide stem is shown in two possible orientations, either side of the glycan backbone, interacting either with G11/K16 or with L38.