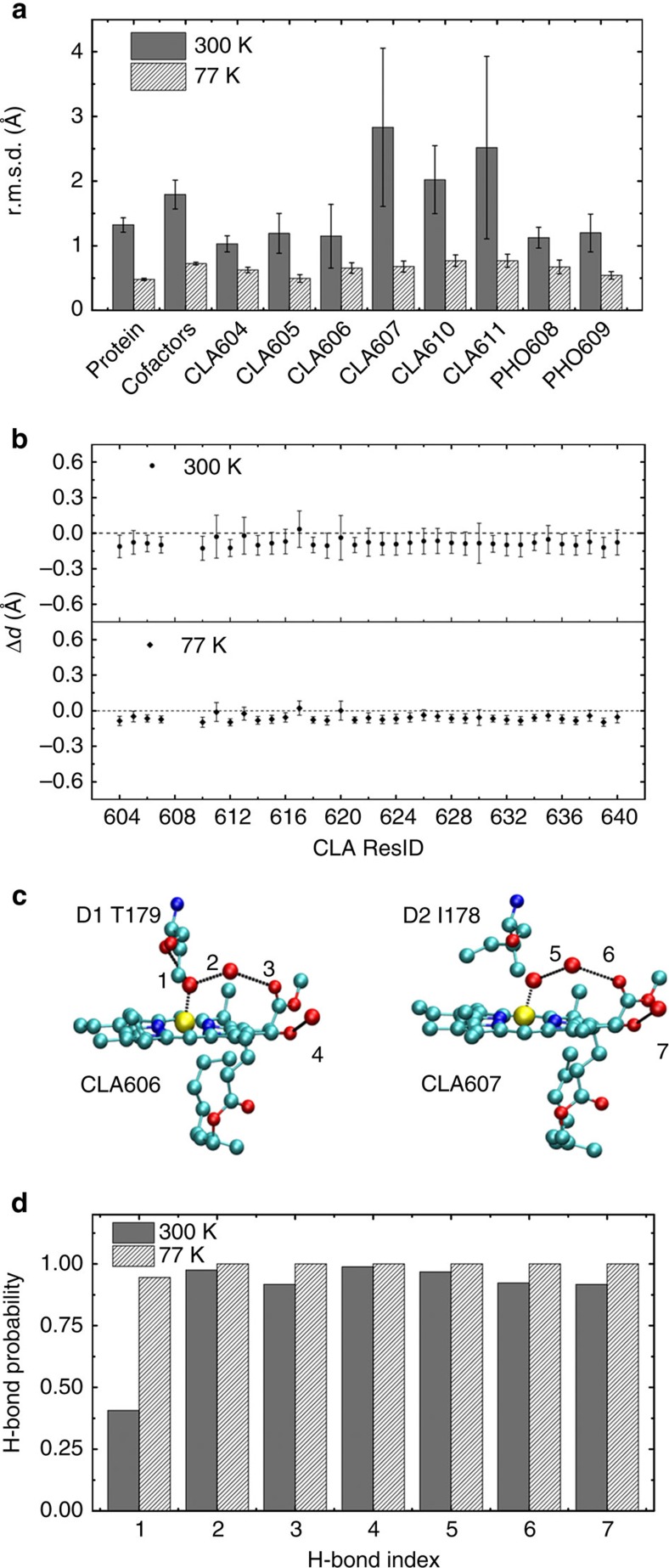
Figure 2. MD simulations preserve important protein–cofactor interactions.
(a) r.m.s.d. with respect to the crystal structure (PDB ID: 3ARC) for various components of the system are calculated based on MD simulations at 300 and 77 K. The r.m.s.d. of the protein and cofactors (located in the electron transfer chains) are computed based on Cα and heavy atoms, respectively. (b) The deviation from the crystal structure of the distance between the Mg in CLA and its coordinated atom. (c) Hydrogen bonds that CLA606 and CLA607 can form with the protein residues in the crystal structure. (d) The fraction of time that an individual hydrogen bond as shown in c is formed in the MD simulations.