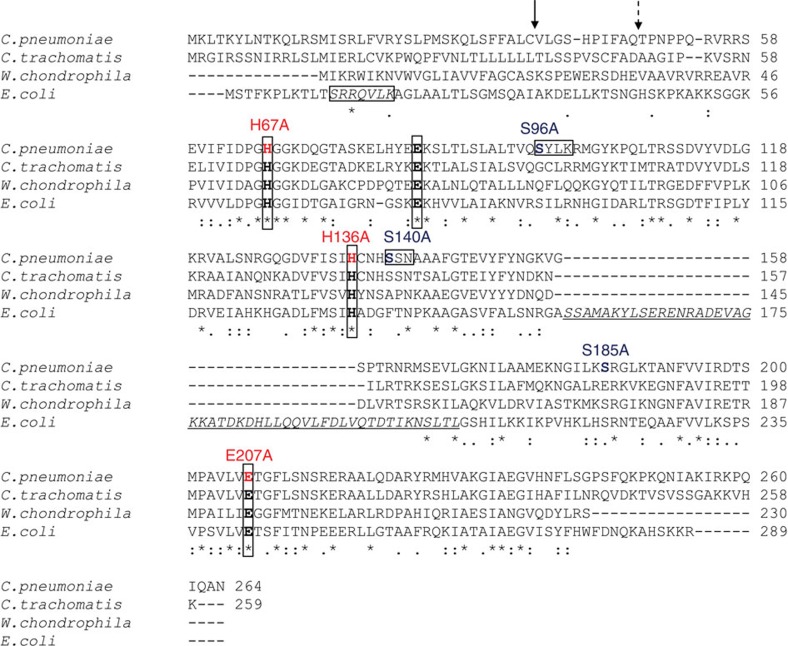
Figure 3. Sequence alignment of AmiA from E. coli and selected Chlamydiales species.
The amidase active site of E. coli AmiA contains three zinc-coordinating residues (H65, E80 and H133), and E242 that is predicted to serve as a general base catalyst9. These active site residues (in bold and boxed) are highly conserved in the Chlamydiales species. Of note, the Chlamydiales AmiA proteins lack a domain with predicted autoregulatory functions (underlined and in italics) that is exclusively found in cell division amidases AmiA, AmiB and AmiC and contains an α-helix (S157-V173 in E. coli AmiA, corresponding to D280-L296 in AmiB from B. henselae) occluding the active site. During cell division, the regulatory domain is proposed to interact with LytM domain factors to relieve autoinhibition by a conformational switch25. PBP DD-CPases have three typical motifs, SxxK, S(Y)xN and K(H,R)T(S)G, that are essential for substrate recognition and catalysis. AmiA from C. pneumoniae contains an SxxK and an SxN motif but lacks the KTG motif. The signal peptide from E. coli contains an SRRxFLK (with x being a polar amino acid) consensus motif (boxed and in italics) that directs AmiA to the Tat translocation system. Consistent with the absence of a Tat system in chlamydiae, AmiA from C. pneumoniae does not contain a functional Tat motif. The solid arrow and the dashed arrow point to the signal peptidase cleavage sites in AmiA from C. pneumoniae revealed by MS analysis and predicted by SignalP32, respectively.