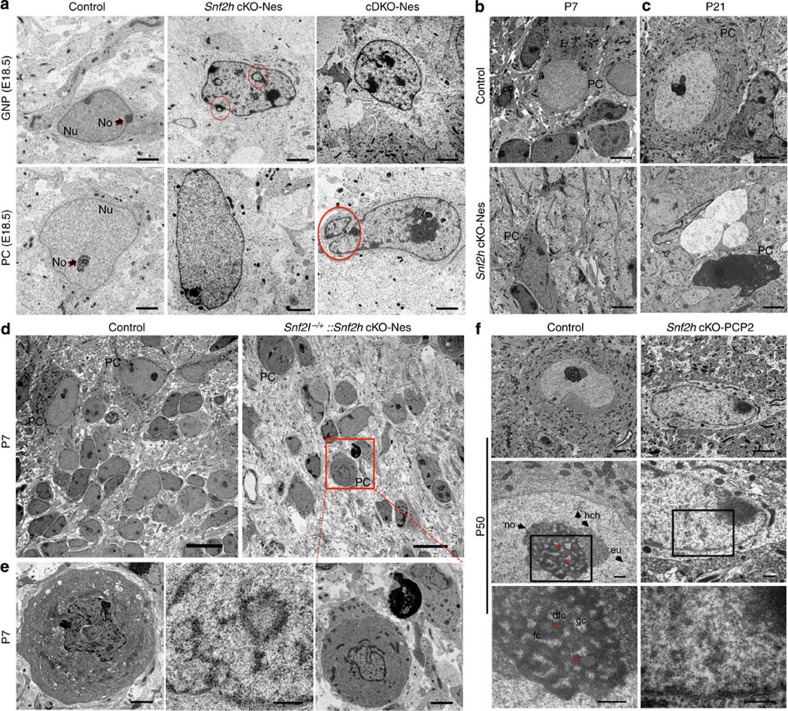
Figure 8. Snf2h is necessary for proper chromatin folding.
(a) Transmission electron microscopy (TEM) of GNPs and PCs through the cerebellar vermis from Snf2h cKO-Nes, cDKO-Nes mice and control mice at E18.5. Red circles denote nuclear invaginations. GNP, granule neuron progenitor; PC, Purkinje cell; Nu, nucleus; No, nucleolus. Scale bars, 2 μm. (b,c) TEM from Snf2h cKO-Nes and control mice through the cerebellum at P7 and P21. Note the abnormal morphology of mutant PCs and ‘cell ghosts’ in mutant cerebella. PC, Purkinje cell. Scale bars, 2 μm. (d,e) TEM through the cerebellar vermis from control and Snf2l−/+::Snf2h cKO-Nes mice at P7, revealing the aggravation of chromatin ultrastructure abnormalities within PCs upon additional removal of one Snf2l copy. Boxed area is enlarged in bottom rightmost panel. Scale bars, 10 μm (top panels); 2 μm (bottom panels). (f) TEM from P50 Snf2h cKO-PCP2 mice and control littermates through the cerebellar vermis. Higher magnification images are provided in descending panels. Note the loss of nucleolar, heterochromatin and euchromatin ultrastructure and increased electron density, evidenced as ‘chromatin clumps’ within mutant PC nuclei. Stars denote nucleolar dense fibrillar centers (dfc). gc, granular centre; fc, fibrillar centre; no, nucleolus; eu, euchromatin; hch, heterochromatin. Scale bars, 2 μm (top panels); 500 nm (middle and bottom panels). At least three mice from each genotype were used for evaluation.