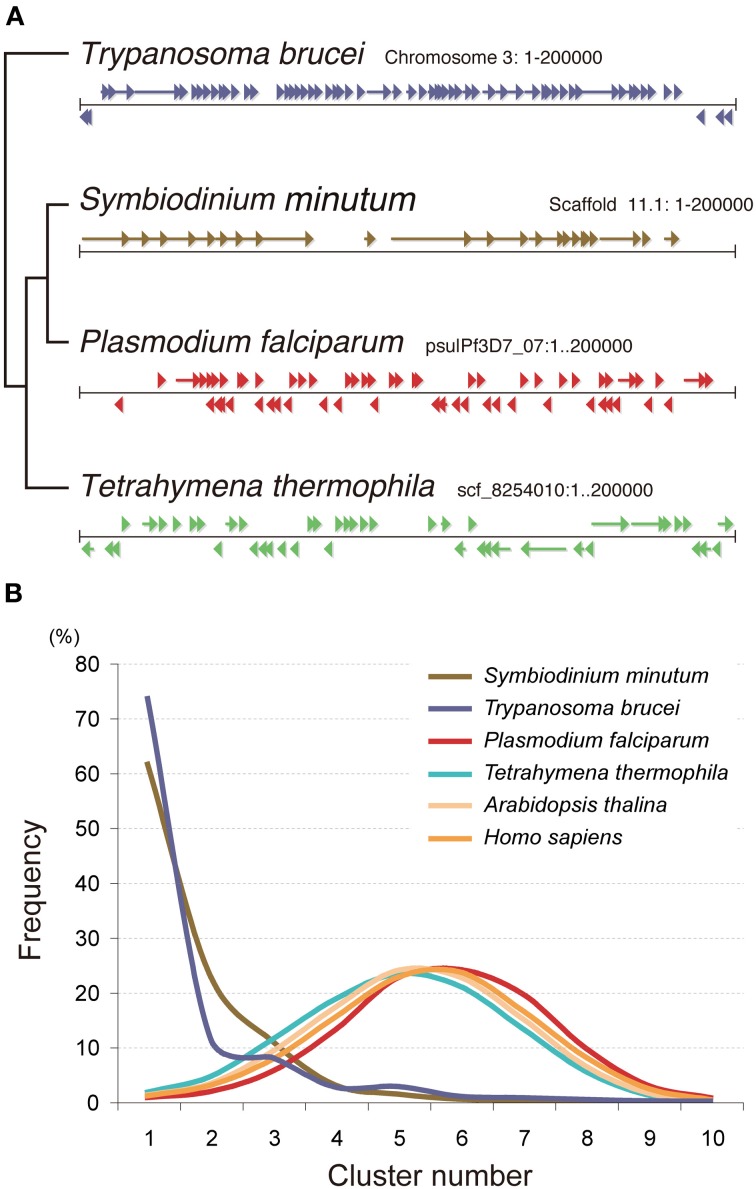
Figure 11.
Nuclear gene arrangement in the Symbiodinium minutum genome. (A) Examples of gene arrangement in the 200-kbp nuclear genome are shown and compared among Symbiodinium minutum (dinoflagellate), Trypanosoma brucei (euglenozoan), Plasmodium falciparum (apicomplexa), and Tetrahymena thermophila (ciliate). In contrast to the genomes of Tetrahymena, Plasmodium, and Acropora, that show a random arrangement of protein-coding genes (arrowheads and arrows), the genomes of S. minutum and Trypanosoma are arranged into a large directional gene cluster in a head-to-tail orientation. (B) A search of the directional gene cluster using a 10-gene window shows the strong tendency toward unidirectional alignment of genes in the S. minutum and Trypanosoma genomes. Each line represents a frequency histogram for changes in the gene orientation between successive genes in the genome. The X-axis represents the number of orientation changes as one moves through windows of 10 genes. For examples as indicating random orientation, the poisson distributions with μ = 4.5 (average) and 0.2 are shown.