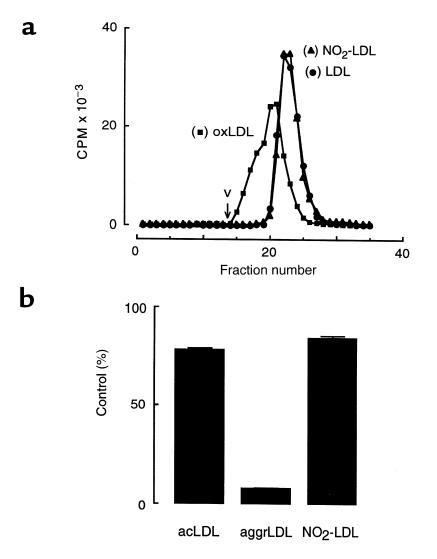
Figure 8.
(a) Size exclusion chromatography. (b) The effect of cytochalasin D treatment on macrophage degradation of modified forms of LDL. (a) Native [125I]LDL (LDL; filled circles), copper oxidized [125I]LDL (oxLDL; filled squares), and [125I]LDL modified by the complete MPO-H2O2-NO2– system (NO2-LDL; filled triangles) were prepared and individually fractionated on a Sephacryl S400-HR column as described in Methods. The elution profiles of native and modified lipoproteins are shown. The void volume (v) of the column is indicated. (b) Acetylated [125I]LDL (acLDL), vortex-aggregated [125I]LDL (aggrLDL), and [125I]LDL modified by the complete MPO-H2O2-NO2– system (NO2-LDL) were prepared as described in Methods. The 125I-labeled lipoprotein preparations were then individually incubated (5 μg/mL) with thioglycollate-elicited MPMs at 37°C for 5 hours in the presence or absence of cytochalasin D (1 μg/mL) in media supplemented with catalase (300 nM) and BHT (20 μM). Cellular degradation of lipoprotein was subsequently determined as described in Methods. Results are expressed as the percentage of lipoprotein degradation observed in the presence vs. absence of cytochalasin D treatment. Lipoprotein degradation by non-cytochalasin D–treated macrophages (control) exposed to acLDL, aggrLDL, and NO2-LDL preparations was 3.69 ± 0.14, 1.78 ± 0.03, and 1.49 ± 0.08 μg LDL per milligram of cell protein, respectively. Data represent the mean ± SD of triplicate determinations from a representative experiment performed in duplicate.