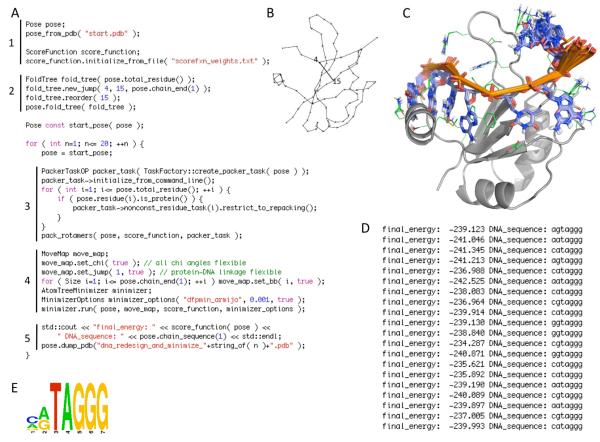
Figure 4.
Simple Rosetta3 protocol for performing a binding specificity calculation on a protein-single-stranded-DNA complex. The simulation code (A) is broken into 5 segments: (1) initialization of the molecular system from a PDB file and the scoring function from a text file containing the energy terms and weights; (2) setup of the kinematic connectivity via a FoldTree (illustrated in B) with a long-range rigid-body connection between residue 4 in the DNA and residue 15 in the protein; (3) redesign of the DNA sequence and simultaneous optimization of the protein sidechain conformations using a PackerTask object to direct the operation of Rosetta’s packing subroutine pack_rotamers; (4) gradient-based minimization of the resulting Pose with flexibility of all chi angles (including glycosidic dihedrals in the DNA), the rigid-body linkage between the protein and the DNA, and the DNA backbone dihedrals (the MoveMap object communicates the allowed flexibility to the minimizer); (5) output of the final optimized structures (superimposed in C) and sequence and score information (text output shown in D, sequences summarized by a sequence logo representation in E, which can be compared with the DNA sequence in the starting PDB file: GTTAGGG). This simulation code could be compiled into a free-standing C++ executable by linking against the Rosetta libraries.