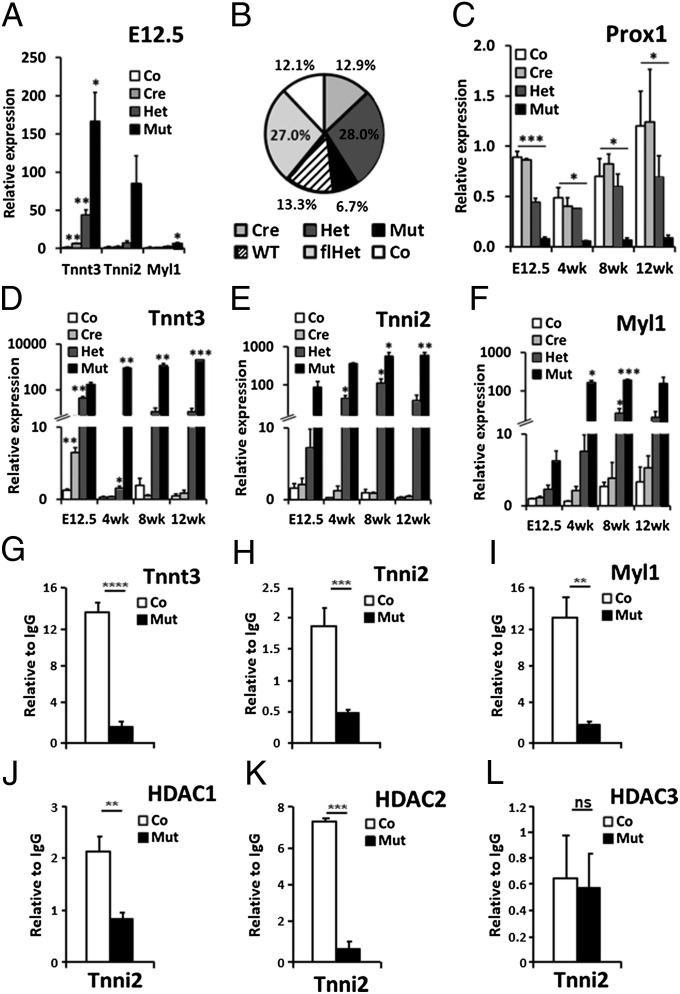
Fig. 1.
Direct regulation of fast-twitch skeletal muscle genes by Prox1 and HDACs. (A) qRT-PCR analysis of Tnnt3, Tnni2, and Myl1 expression in control (Co, Prox1fl/fl), cre (Cre, Nkx2.5Cre/+;Prox1+/+), heterozygous (Het, Nkx2.5Cre/+;Prox1fl/+), and mutant (Mut, Nkx2.5Cre/+;Prox1fl/fl) E12.5 hearts (n = 3) to confirm microarray findings (SI Appendix, Table S1). (B) Ratio of genotypes at P10 from Nkx2.5Cre/+;Prox1fl/+ × Prox1fl/+ cross (n = 556) deviated significantly from expected Mendelian inheritance (P = 0.002). WT, Prox1+/+; flhet, Prox1fl/+. (C) Prox1 expression in control, cre, het, and mutant hearts at E12.5 (n = 3) and 4 (n = 2), 8 (n = 3), and 12 wk (n = 3) expressed relative to an E12.5 control heart. Significance determined by Student t test in comparison with control levels at each stage: *P ≤ 0.05; **P ≤ 0.01; ***P = ≤ 0.001. Analysis of control, cre, het, and mutant hearts to demonstrate increased expression of Tnnt3 (D), Tnni2 (E), and Myl1 (F) in mutant hearts at E12.5, and 4, 8, and 12 wk. Relative expression, N numbers, and significance are as in C. ChIP–qPCR quantification of Prox1 (G–I), HDAC1 (J), HDAC2 (K), and HDAC3 (L) antibody-immunoprecipitated chromatin from adult hearts of control (Prox1fl/fl; n = 6; white bars) and mutant (Nkx2.5Cre/+;Prox1fl/fl; n = 3; black bars) hearts using primers targeting responsive elements within Tnni2 (A and J–L), Tnnt3 (H), and Myl1 (I) genomic loci. ChIP–qPCR signals were standardized to the percentage of input DNA and normalized to IgG ChIP signals to determine the relative enrichment of anti-Prox1, HDAC1, HDAC2, or HDAC3 antibody over IgG immunoprecipitated chromatin. Each ChIP–qPCR was done in triplicate. Loss of binding in mutant hearts determined by Student t test comparison with control heart chromatin: ns, not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.