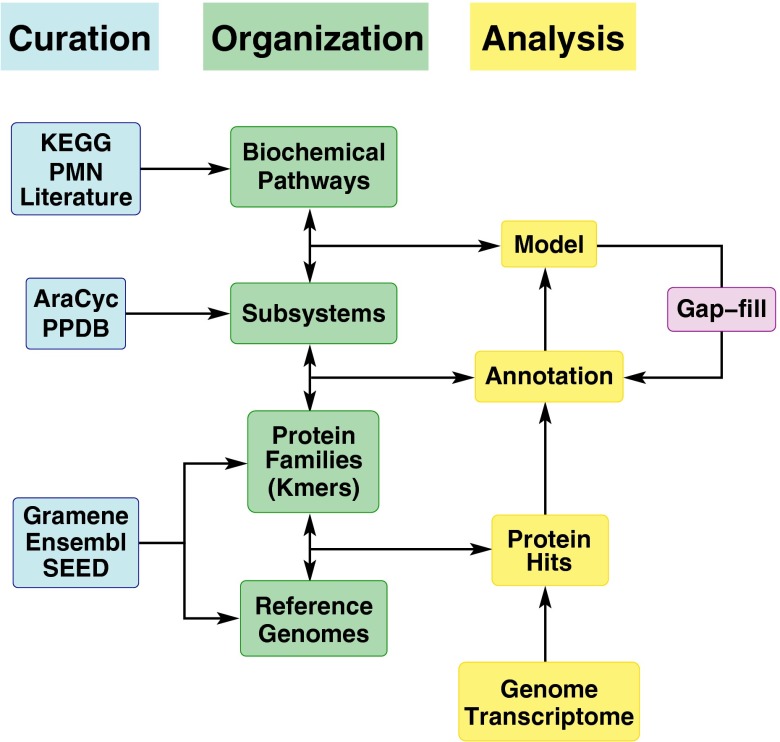
Fig. 1.
Overview of PlantSEED, which consists of reference genomes, protein families, subsystems, and biochemical pathways. Reference genomes for the 10 species listed in Table 1 were installed from Gramene. Protein families computed by Ensembl Compara for these 10 genomes were installed along with Kmers (unique oligopeptide sequences representing the families) computed by SEED. Gene–reaction associations were curated primarily using AraCyc metabolic pathways to form a set of PlantSEED subsystems. Finally, a biochemical pathway database was formed by integrating KEGG, several BioCyc databases from PMN and Gramene, and published metabolic models. Gapfilling results from use of the reference genomes are reviewed and used in PlantSEED’s subsystems and resulting annotation, thereby improving PlantSEED’s output dynamically.