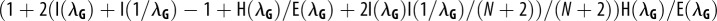Table 1.
Overview of published indices related to the concept of integration. In the definitions, N is the number of traits, λ is the eigenvalues of the correlation matrix, r is the set of pairwise correlation coefficients, E denotes the expectation (the average) and |x| denotes the absolute value of x.
| index | notes | references |
|---|---|---|
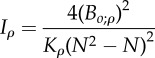 |
a complex index related to the fraction of correlations above a fixed threshold, and scaled to lie between 0 and 1a | Olson & Miller [5] |
| Iz = tanh(E(|z|)) | average of Fisher's z-transformed correlation back transformed to 0–1 scaleb | Van Valen [32] |
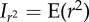 |
average coefficient of determination, estimated as the mean of the squared pairwise correlations | Van Valen [159] |
| Ir = E(|r|) | average of the absolute pairwise correlations | Cane [160] |
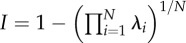 |
one minus the geometric mean of the correlation-matrix eigenvalues | Cheverud et al. [161] |
| var(λ) | variance of the correlation-matrix eigenvalues | Wagner [162], Cheverud et al. [163] |
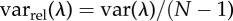 |
relative variance of the correlation-matrix eigenvalues; the value N − 1 is the maximum possible variance of an eigenvalue | Pavlicev et al. [164] |
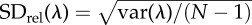 |
relative standard deviation of the correlation-matrix eigenvalues | Cheverud et al. [161], Pavlicev et al. [164] |
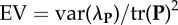 |
the variance of the variance-matrix eigenvalues (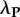 ) scaled by the total variancec ) scaled by the total variancec
|
Young [165], Willmore et al. [166] |
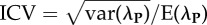 |
the standard deviation of the variance-matrix eigenvalues scaled by the mean of the eigenvalues | Shirai & Marroig [167] |
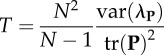 |
one of Van Valen's [159] measures of ‘tightness’, the closeness of the distribution to the major axis, varying between 0 and 1 | Van Valen [159] |
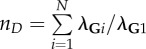 |
the sum of the genetic variance matrix eigenvalues (λG) scaled by the leading eigenvalue (λG1) | Kirkpatrick [168] |
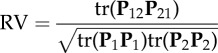 |
a measure of the total amount of covariation between the two sets of variables over a measure of the total amount of variation in the within the two groupsd | Klingenberg [98] |
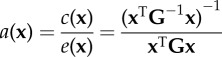 |
the fraction of independent additive genetic variation (autonomy) for a particular linear combination of the traits (x)e | Hansen & Houle [169] |
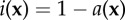 |
the fraction of non-independent additive genetic variation (integration) in the direction of x | Hansen & Houle [169] |
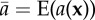 |
the average autonomy of uniformly distributed random directionsf | Hansen & Houle [169] |
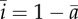 |
the average integration of uniformly distributed random directions | Hansen & Houle [169] |
aBo;ρ is number of correlations above, or equal to, the lower statistical significance level (a function of sample size) of a fixed arbitrary threshold correlation given by ρ. Kρ is the number of non-contained ρ-groups, where non-contained means the largest group which can be formed where all elements have pairwise correlations ≥ ρ.
btanh is the inverse Fisher transformation (the hyperbolic tangent), z is a set of Fisher's z-transformed pairwise correlation coefficients.
ctr is the trace function. P is the phenotypic variance matrix.
dP1, P2, P12 and P21 are the sub matrices of a phenotypic variance matrix. The sub matrices P1 and P2 are the variance matrices for the two sets of traits, respectively. The sub matrices P12 and P21 are the covariances between the two sets of traits.
ee(x) is the evolvability and c(x) the conditional evolvability along a unit length vector (or direction) of the traits x, G is the additive genetic covariance matrix, T denotes the transpose, and −1 denotes the inverse. To calculate the autonomy to a specific trait with respect to the rest, we can use a vector x with the coefficient 1 for the focal trait and zero for the rest. The different indices from Hansen & Houle [169] are easily computed using the R package ‘evolvability’ (see [156]).