Abstract
Unveiling of cancer genomes is unleashing new therapeutic strategies for cancer. With cancer parts lists in hand, new approaches to personalized medicine can be developed by understanding the assembly of cancer machines using modular domains in proteins and their associated networks. Using the Src-homology-2 (SH2) domain as an example, new profiling approaches can discern global patterns of tyrosine phosphorylation in cancer cells that can enable molecular cancer medicine. Identifying and quantifying protein–protein interactions also has the potential to subtype tumors and guide clinical decision making. These approaches should extend the impact of genomics through viewing the architecture of cancer systems and improve predictions of patient outcome and therapeutic response, as well as guide combination therapy approaches that attack cancer systems.
Keywords: Cancer, Domain, SH2, Tyrosine kinase, Personalized medicine, Kinase inhibitor
1. Personalized cancer medicine
This perspective is written from the viewpoint of a practicing oncologist interested in applying protein modules and networks toward new diagnostic and therapeutic approaches to cancer. We have seen improvements in cancer care over the last decade produced in large part by a better understanding of the genomic mechanisms defining cancers [1]. The overarching goal of modern cancer medicine is to identify tumor subsets that depend on particular targets for survival and direct therapy against these disease-specific drivers of cancer. The ability of practicing clinicians to match therapy with an individual patient has the potential to dramatically improve survival with reduced toxicity. For example, major progress has been made against subsets of cancer using inhibitors of tyrosine kinase signaling molecules as a therapeutic strategy. Perhaps the best example is the use of imatinib (Gleevec) for BCR-ABL dependent chronic myelogenous leukemia (CML) [2]. The success of imatinib was heavily influenced by the knowledge of the BCR-ABL tyrosine kinase and its critical importance to leukemia cells. Similar stories include the use of imatinib for gastrointestinal stromal tumors (GIST), herceptin for HER2 overexpressing breast cancer, gefitinib/erlotinib for epidermal growth factor receptor (EGFR) mutant driven lung cancers, and crizotinib for EML4-ALK driven lung cancers [3-6]. Routine molecular testing for EGFR mutations or EML4-ALK rearrangements, now a standard practice at our institution and others, have led to personalized medicine approaches using small molecule tyrosine kinase inhibitors (TKI) to elicit responses and improve outcome for lung cancer patients [7].
Despite this success using genomic based approaches, a number of limits exist. Approximately 10% of patients with advanced adenocarcinoma of the lung have activating EGFR mutations and other tumor types may be lacking classic actionable driver oncogenes. Tumor responses, both in terms of degree of tumor shrinkage and time of response, can be heterogeneous. Tumors having a common oncogene driver (i.e. EGFR mutation) can display differences in response to TKI, such that degree of tumor shrinkage and time of response can vary from patient to patient [8,9]. In solid tumors such as lung and breast cancer, the responses can be short lived with rapid emergence of resistance and regrowth of the tumor. Response heterogeneity has been explained in part by differences in basal pre-treatment levels of pro-apoptotic proteins, such as BIM, as well as quantitative differences in pre-existing resistant cells. Genetic heterogeneity shaped by selection pressures is being increasingly recognized as a critical factor in carcinogenesis and next-generation DNA sequencing platforms are starting to describe this diversity landscape [10,11]. For example, Su found that higher levels of pre-treatment gatekeeper mutant EGFR alleles (T790M) that produce EGFR TKI insensitive proteins predicted shorter response duration in patients treated with EGFR TKI [12]. Finally, in addition to increasingly complex biology, questions are beginning to be raised about the true benefit of targeted therapy and the costs associated with the therapy [13].
Here I will argue that, while major advances have been enabled by genomic medicine, the next major leap will occur in understanding how cancer machine are built from modular domains and assembled into networks. There is little doubt that genome based approaches will expand in the future of personalized cancer medicine. However, an overlapping wave of progress could further augment the genomic approach by applying lessons learned from modular domain proteins and their assembly into disease related networks. Such a view can further identify subsets of cancers not fully articulated by genomics, can define new therapeutic targets, and can start to enable rationale combinations of drugs in an individualized fashion.
2. Network medicine
There is increasing recognition that biological networks play an important role in cancer biology. Rather than single proteins working in isolation or simple unidirectional pathways, signaling proteins in cancer behave more as networks, with dense interconnections, multifunctional roles of proteins, reliance on protein complexes to elicit molecular function, and robustness [14-16]. Cancer is recognized to be a result of changes in cellular genomes resulting in aberrant signaling proteins causing deregulated cell growth, survival, and metastasis. These changes rewire entire signaling ‘circuits’ resulting in aberrant growth and metastasis. Critical to protein function and signaling is the formation of signaling complexes and networks of signaling proteins that act in concert to produce a physiological signal [17,18]. State of the art mass spectrometry is now able to accurately map protein–protein interaction (PPI) complexes and signaling networks using phosphoproteomics[19,20]. This now allows for a more global view of how cancer proteins drive a signaling network to transform cells. The application of network theory to biology also enables a better understanding of cancer, improve ability to classify tumors, and suggest therapeutic approaches against cancer ‘hub’ proteins or suggest rational combination approaches [14-16,21,22]. In addition, comprehensive PPI databases (such as [23]. Cui and colleagues used cancer genome data to produce oncogene networks that appear in solid tumors including breast and lung cancer and such networks could simplify mutational data when viewed through a more global approach [24]. Thus, how to make ‘network medicine’ become a reality [25]?
3. Modules and networks as diagnostics
Early studies using gene expression profiling raised awareness that similarly appearing tumors nonetheless could display marked differences when viewed through the lens of molecular profiling tools [26,27]. Likewise, tumors profiled using mass spectrometry based approaches demonstrate similar properties. For example, mass spectrometry (MS) coupled with anti-phosphotyrosine (anti-pTyr) antibodies to profile tyrosine kinase signaling revealed different patterns of tyrosine kinase signaling in lung cancer cells and tumors [28]. Overall, this work provided proof of principle that characterization of global tyrosine phosphorylation patterns can provide useful information for classifying lung cancers. The importance of examining global tyrosine phosphorylation is especially relevant as recent studies have found enrichment of phosphopro-teins encoded by disease-associated genes and disease seems to rely on more common evolutionarily conserved networks, including cancer that relies on pTyr networks [29,30].
One of the most important consequences of protein tyrosine phosphorylation is to regulate protein-protein interactions [31]. Many tyrosine-phosphorylated proteins serve as high-affinity binding sites for proteins containing modular pTyr-specific binding domains. These modular domains serve to couple tyrosine phosphorylation to the assembly of signaling complexes and the relocalization of signaling proteins, and thus play a central role in downstream signaling. By far the most abundant module in humans is the Src Homology 2 or SH2 domain [32]. There are 120 SH2 domains encoded by the human genome [33,34], and each SH2 domain has binding specificity for a unique spectrum of tyrosine phosphorylated sites [35]. Because SH2 domains are what the cell actually uses to respond to or “read” changes in tyrosine phosphorylation “written” by tyrosine kinases, the extent of binding of different SH2 domains to a cell sample can provide a great deal of information about the signaling state and its underlying mechanisms. Thus, this approach could be useful in characterizing and classifying complex tumor types, especially cancers where multiple tyrosine kinases can be driving downstream signaling pathways and maintaining tumor growth [1].
Bruce Mayer and his group developed a novel phosphoproteomic method termed SH2 profiling, to profile phosphotyrosine (pTyr) signaling in cancer cells [35-38]. The conceptual basis of SH2 profiling is to take advantage of the cell’s own pTyr binding motifs and use them as probes for the state of tyrosine phosphorylation of a cell sample. This approach can discern differences in SH2 profiles (and therefore pTyr signaling) in cells transformed by distinct oncogenic tyrosine kinases [35], and could classify subtypes of leukemia [36]. Together our two groups recently reported how SH2 profiling can provide information on global tyrosine kinase signaling in lung cancers and how this may be useful for future personalized medicine approaches [39]. SH2 domain profiles were generated using a panel of purified SH2 domains on multiple lung cancer cell lines that harbor different oncogene mutations (EGFR, KRAS) as well as display differences in drug sensitivity to EGFR tyrosine kinase inhibitors. When SH2 domain profiles were examined using unsupervised clustering, we could observe groups of cells based on SH2 binding patterns with some clusters correlated with EGFR or KRAS mutation status. Amongst cells displaying identical histology and presence of activating EGFR mutations, we could observe striking differences in global tyrosine kinase signaling patterns amongst cells in this group, suggesting the added value of SH2 domains in defining global tyrosine kinase signaling in cells driven by a common oncogene. For example, cells with hyperactivated EGFR through mutation could in some instances cluster amongst cells lacking EGFR mutations. Conversely, we observed some cell lines that, despite having non-mutated EGFR, had SH2 profiles indicative of hyperactive EGFR signaling. We also observed cells with hyperactivated MET signaling displaying unique patterns of SH2 binding, suggesting the ability of using modular domain profiling with SH2 domains to read out distinct upstream tyrosine kinase signaling molecules. Furthermore, a set of SH2 probe binding correlated with the sensitivity of the cells to EGFR tyrosine kinase inhibitors, suggesting these domains could serve as predictive biomarkers for TKI therapy. Lastly, SH2 domain binding could be perturbed in cancer cells using tyrosine kinase inhibitors and we could observe similarities in perturbations between two distinct inhibitors, suggesting overlap of critical targets. Overall this study illustrates the potential of SH2 domain profiles as molecular diagnostics to both classify tumors and define upstream tyrosine kinases vulnerable to attack.
Currently, we are attempting to classify lung cancers taken directly from patients on the basis of quantitative SH2 profile results and attempt to correlate with clinical outcomes. We are interested in identifying patterns of SH2 binding that correlate with clinically relevant properties of lung cancers, such as EGFR mutation, pathologic features such as histology and stage, and with clinical outcomes (survival). Domain profiling using SH2 domains could also serve as predictive biomarkers for kinase inhibitor therapy. By virtue of their ability to read patterns of tyrosine phosphorylation driven by tyrosine kinases, patterns of SH2 binding may not simply produce a proxy for genetic oncogene addiction but, possibly more importantly, can signal the presence of multiple tyrosine kinases cooperating in driving tumor growth and survival. Expansion of this approach to improve feasibility in hospital diagnostic laboratories with streamlined quantitative analysis could allow for wide scale profiling of cancers. Finally, dynamic responses of SH2 domains, especially in the face of kinase inhibitors, may help define not only acquired resistance to TKI therapy but also intrinsic resistance. Most reported mechanisms underlying resistance to TKI therapy result from genetic lesions, such as secondary mutations or gene amplification of other receptor tyrosine kinases that crosstalk to important signaling pathways [40]. SH2 domains change binding patterns in response to TKI and in some instances we have observed increased SH2 binding despite inhibition of tyrosine kinase signaling with TKI. It remains to be determined if SH2 domains facilitate formation of new complexes in the face of TKI therapy and if this is one way for tumors to survive TKI therapy. While activation of other kinases (including those with SH2 domains) is frequent in drug resistance, little attention has been paid to the ability of non-catalytic adaptor proteins with modular domains such as SH2 that can form new complexes following TKI therapy. CRK-L, for example, has recently been implicated as a non-kinase SH2 containing adaptor program that can mediate TKI resistance in lung cancer cells addicted to EGFR [41].
4. Translating interactomes into molecular profiles
Encouraged by the results using SH2 domains that identify distinct patterns of pTyr within a common oncogene genotype (EGFR), our lab and the Superti-Furga lab are undertaking a system wide and global analysis of the protein-protein interactomes driven by hyperactivated EGFR resulting from somatic mutation in lung cancer [42-44]. Our approach has been to use tandem affinity purification – mass spectrometry (TAP-MS) in cell models that harbor mutant EGFR proteins and are highly sensitive to EGFR TKI. Tagged versions of EGFR are initially used to identify interactions and second round of TAP-MS is performed to more fully construct the network [45]. This interaction network will allow functional interrogation of targets important in driving mutant EGFR growth and survival signals. The interactions themselves could define an addicted EGFR reference map and, similar to observed with SH2 domains, differences in the interactome could further define different subtypes of tumor cells within a common genotype. This could be relevant to clinical observations of different degrees of response to EGFR TKI despite same gentoypes and emergence of resistant cells [8,40]. Thus, the critical question becomes, how can one ‘translate’ these highly sophisticated interactome data, whether generated by mass spectrometry or other approaches, and actually detect and quantify interactomes in human cancer samples?
Translation of these interactome approaches to tumor samples is hampered by a number of hurdles. First, almost all studies have been carried out in engineered mammalian cells that express a tagged version of the protein of interest; this limits the ability to use these modern TAP-MS approaches in samples from patients. Second, most samples from patients in clinical practice are formalin fixed and paraffin embedded. This precludes the ability to use fresh frozen tissue for immunoprecipitation and western blotting (IP-WB) to identify protein complexes. IP-WB also requires a large amount of starting material that can be difficult to obtain with needle biopsies. One solution to mapping networks identified using MS-based proteomics is proximity ligation assays (PLA) [46-51]. Briefly, two proteins in complex are each identified with primary antibodies specific for the protein and linked to a conjugated oligonucleotide. A connector oligonucleotide links both proximity probes allowing ligation and formation of a template for PCR amplification. The resulting rolling circular amplification (RCA) serves as a target for hybridized fluorescently labeled detection oligonucleotides allowing distinct and bright spots to be identified and quantified in a fluorescent microscope. See http://www.olink.-com/movie.php for a graphic movie of this technology. This technology has been used to identify in cells in situ Myc–Max interactions, tyrosine phosphorylation of receptor tyrosine kinases, and interactions of proteins in human tissue sections.
As little has been done to establish biomarker systems to measure protein-protein based biomarkers in cancer, our group is currently developing PLA that can quantitatively measure defined EGFR protein-protein interactions in lung cancer specimens and relate expression of these interactions to clinical outcome variables.
The approach to develop biomarkers based on in situ protein–protein interactions goes one step beyond measurement of protein expression as it determines the binding to two proteins together in tumor tissues. Two proteins may be equally expressed, but because of other nuances in the cancer cell, do not form a signaling complex that drives a signaling cascade. In another tumor cell, these proteins form a complex and drive a signaling cascade. Gene-based approaches that measure mRNA expression or immunohistochemistry approaches that measure protein expression would be unable to discriminate these two cases. Conceptually, we can build on our interactome maps, either directly generated through mass spectrometry based experiments or through databases data, to construct in situ protein–protein interaction networks in patient based materials. This may lead to further subtyping a cancer despite common genotype and mark interactions as targets for future chemistry.
5. Challenges
It is becoming more apparent that cancers, especially solid tumors such as lung cancer, are the result of multi-gene hits resulting from environmental insults coupled with genetic predispositions to development of cancer. This includes gain or loss of multiple chromosomal elements within single tumors as well as multiple mutations in cancer causing genes such as kinases [52-54]. It is clear that genotypes will be a common diagnostic tool for practicing physicians yet important nuances of cancers could still be realized through views of how modular domains organize in tumor cells. One of the challenges is to understand how all these genetic changes ultimately converge on growth, survival, and metastasis signaling programs to allow the development of cancer. Here network analysis approaches can help integrate various genomic data to produce cohesive views of deregulated pathways affecting patient outcome [23,55]. A further challenge is to integrate these changes to define novel therapeutic opportunities for patients with cancer. One of the limits of current cancer medicine is the lack of rationale combinations of molecularly targeted agents. One can view progress in other diseases, especially infectious diseases, to get clues about how to make progress in cancer. For example, management of HIV progressed from single agent antiretroviral therapy with AZT to multi-agent antiviral therapy (HAART) [56]. Therefore, combination approaches against a system can be highly effective in treating complex diseases. Translation of the underlying science of how cancer proteomes are assembled using their modular domains could be one way forward towards rationale combinations of pathway inhibitors designed and personalized for each individual’s cancer (Fig. 1). In the near future it is likely to have the entire cancer parts list at the fingers for oncologists, yet assembly of these enormous datasets into a cohesive and integrated cancer system that predicts vulnerability will limit efficacy of such datasets. Down-sides of such an approach is the logistical hurdles of combination therapy related to regulatory development of compounds, economic cost, and potential for serious toxicity when manipulating multiple signaling proteins in normal tissues. Better predictions of the activity of therapeutics is critical, as testing multiple compounds and combinations in patients with cancer is prohibitive for a number of reasons, such as limited financial resources, difficulties with combinations of unapproved agents, and low enrollment on clinical trials [57,58]. As we have discussed here, the biology of modular domains on a system wide level could tease out differences that could be important from a prognostic or therapeutic vantage point [59]. Towards this end, systems medicine teams need to be created and supported to handle the multidimensional data and provide enough statistical support for biomarker development.
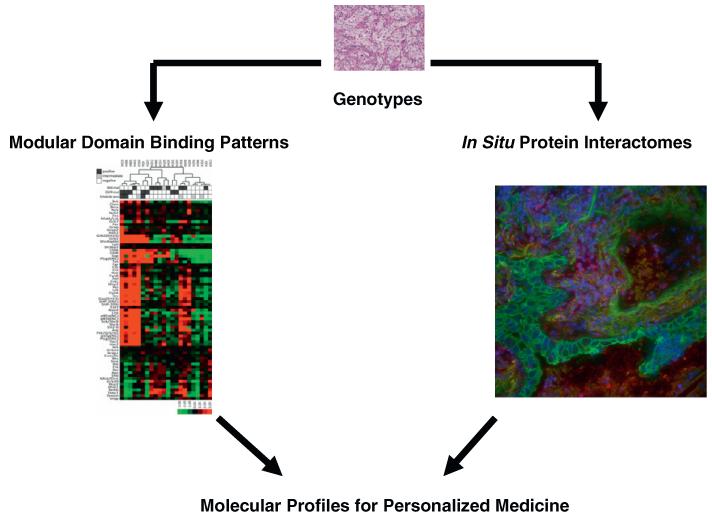
Fig. 1.
A role for modular domain binding patterns and interactome networks in molecular medicine. Tumor tissues will receive genotype analysis to examine from driver mutations in oncogenes and/or examine for loss of tumor suppressor genes that represent different subtypes of tumors based on genotypes. Modular domain binding patterns, for example SH2 domains that recognize phosphotyrosine, can produce additional views of tumors. In the case demonstrated here, SH2 domain profiling can identify a strong pattern of SH2 binding in tumor cells (red). Similarly, pre-defined protein–protein interactions measured using proximity ligation assays can identify tumor interactomes that could further classify tumors. On the right, red foci indicate interactions between EGFR and Grb2 proteins measured using proximity ligation assays, nuclei are identified with DAPI (blue) and tumor cells identified using cytokeratin (green). Development of these approaches into validated biomarker systems could classify tumors and provide information relevant for clinical care, including prognostic measures as well as predictive biomarkers for drug therapy. SH2 domain figure is shown as published in Ref. [39]. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Lastly, will we see new attempts to disrupt signaling using drugs acting as disruptors of protein–protein interactions or remodelers of signaling using principles of synthetic biology? [60] One of the larger drug targets has been kinases and this has led to many targets being developed with large amounts of academic and industry overlap [61]. However, most solid tumors addicted to oncogenes demonstrate resistance mechanisms that can be hard to overcome with subsequent therapeutics. This raises the possibility of using protein–protein interaction disrupters to produce additional pressure on tumor cells to escape drugs targeting their addiction mechanisms [62]. Recent reviews highlight emerging approaches to develop compounds disrupting protein–protein interactions and early results of compounds in human clinical trials, such as navitoclax and obatoclax that disrupt Bcl2 family complexes, have been reported [63-65]. One could also hope to see new therapeutics built upon synthetic biology that promote rewiring modular domains and networks to drive tumor cells back towards a normal state.
Acknowledgements
I thank members of the Moffitt ‘pY Proteomics’ group for helpful discussions, Matthew Smith for the PLA images, Kazuya Machida and Bruce Mayer at the University of Connecticut for fruitful collaborations, and Giulio Superti-Furga and colleagues at CeMM for hosting my sabbatical and for helpful discussions. Work in my laboratory is funded by grants from the National Institute of Health (Moffitt Lung Cancer SPORE P50-CA119997, 1R01CA121182-01A1, RC1CA146843, R21CA162178) and the National Functional Genomics Center (W81XWH-08-2-0101).
References
- [1].Sledge GW., Jr. The challenge and promise of the genomic era. J. Clin. Oncol. 2012;30:203–209. doi: 10.1200/JCO.2011.39.0088. http://dx.doi.org/10.1200/JCO.2011.39.0088. [DOI] [PubMed] [Google Scholar]
- [2].Druker BJ, David A. Karnofsky Award lecture. Imatinib as a paradigm of targeted therapies. J. Clin. Oncol. 2003;21:239s–245s. doi: 10.1200/JCO.2003.10.589. [DOI] [PubMed] [Google Scholar]
- [3].Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- [4].Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- [5].Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat. Med. 2012;18:349–351. doi: 10.1038/nm.2697. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- [6].Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, Massion PP, Siwak-Tapp C, Gonzalez A, Fang R, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 10:760–774. doi: 10.1038/nrc2947. doi: nrc2947[pii]10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, Zakowski MF, Kris MG, Ladanyi M, Miller VA. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. doi: 12/3/839[pii]10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- [9].Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat. Rev. Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- [11].Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci. Transl. Med. 2012;4:127ps110. doi: 10.1126/scitranslmed.3003854. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- [12].Su KY, Chen HY, Li KC, Kuo ML, Yang JC, Chan WK, Ho BC, Chang GC, Shih JY, Yu SL, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J. Clin. Oncol. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- [13].Fojo T, Parkinson DR. Biologically targeted cancer therapy and marginal benefits: are we making too much of too little or are we achieving too little by giving too much? Clin Cancer Res. 16:5972–5980. doi: 10.1158/1078-0432.CCR-10-1277. doi: 16/24/5972 [pii] 10.1158/1078-0432.CCR-10-1277. [DOI] [PubMed] [Google Scholar]
- [14].Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. doi: 10.1038/ nrg1272 nrg1272 [pii] [DOI] [PubMed] [Google Scholar]
- [15].Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc. Natl. Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. doi: 0701361104 [pii] 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. doi: nbt1338 [pii] 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- [17].Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- [18].Faber AC, Li D, Song Y, Liang MC, Yeap BY, Bronson RT, Lifshits E, Chen Z, Maira SM, Garcia-Echeverria C, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc. Natl. Acad. Sci. USA. 2009;106:19503–19508. doi: 10.1073/pnas.0905056106. doi: 0905056106 [pii] 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 2009;5:237. doi: 10.1038/msb.2008.75. doi: msb200875 [pii] 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 2009;10:617–627. doi: 10.1038/nrg2633. doi: nrg2633 [pii] 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- [21].Barabasi AL. Network medicine-from obesity to the “diseasome”. N. Engl. J. Med. 2007;357:404–407. doi: 10.1056/NEJMe078114. doi: NEJMe078114 [pii] 10.1056/NEJMe078114. [DOI] [PubMed] [Google Scholar]
- [22].Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. doi: S0092-8674(06)00439-9 [pii] 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- [23].Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. doi: nbt.1522 [pii] 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- [24].Cui Q, Ma Y, Jaramillo M, Bari H, Awan A, Yang S, Zhang S, Liu L, Lu M, O’Connor-McCourt M, et al. A map of human cancer signaling. Mol. Systems Biol. 2007;3:152. doi: 10.1038/msb4100200. doi: 10.1038/msb4100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pawson T, Linding R. Network medicine. FEBS Lett. 2008;582:1266–1270. doi: 10.1016/j.febslet.2008.02.011. doi: S0014-5793(08)00115-4 [pii] 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- [26].Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- [28].Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- [29].Tan CS, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jorgensen C, Bader GD, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2009;2:ra39. doi: 10.1126/scisignal.2000316. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]
- [30].Li L, Tibiche C, Fu C, Kaneko T, Moran MF, Schiller MR, Li SS, Wang E. The human phosphotyrosine signaling network: Evolution and hotspots of hijacking in cancer. Genome Res. 2012 doi: 10.1101/gr.128819.111. doi: 10.1101/gr.128819.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- [32].Machida K, Mayer BJ. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim Biophys Acta Proteins and Proteomics. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]
- [33].Liu BA, Shah E, Jablonowski K, Stergachis A, Engelmann B, Nash PD. The SH2 domain-containing proteins in 21 species establish the provenance and scope of phosphotyrosine signaling in eukaryotes. Sci. Signal. 2011;4:ra83. doi: 10.1126/scisignal.2002105. doi: 10.1126/scisignal.2002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol. Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. http://dx.doi.org/10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- [35].Machida K, Thompson CM, Dierck K, Jablonowski K, Kärkkäinen S, Liu B, Zhang H, Nash PD, Newman DK, Nollau P, et al. High-throughput phosphotyrosine profiling using SH2 domains. Mol. Cell. 2007;26:899–915. doi: 10.1016/j.molcel.2007.05.031. [DOI] [PubMed] [Google Scholar]
- [36].Dierck K, Machida KAV, Thimm J, Horstmann M, Fiedler W, Mayer BJ, Nollau P. Quantitative multiplexed profiling of cellular signaling networks using phosphotyrosine-specific DNA-tagged SH2 domains. Nat. Meth. 2006;3:737–744. doi: 10.1038/nmeth917. [DOI] [PubMed] [Google Scholar]
- [37].Machida K, Mayer BJ, Nollau P. Profiling the global tyrosine phosphorylation state. Mol. Cell. Proteomics. 2003;2:215–233. doi: 10.1074/mcp.R300002-MCP200. [DOI] [PubMed] [Google Scholar]
- [38].Nollau P, Mayer BJ. Profiling the global tyrosine phosphorylation state by Src Homology 2 domain binding. Proc. Natl. Acad. Sci. USA. 2001;98:13531–13536. doi: 10.1073/pnas.241215998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Machida K, Eschrich S, Li J, Bai Y, Koomen J, Mayer BJ, Haura EB. Characterizing tyrosine phosphorylation signaling in lung cancer using SH2 profiling. PLoS ONE. 2010;5:e13470. doi: 10.1371/journal.pone.0013470. http://dx.doi.org/10.1371/journal.pone.0013470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. http://dx.doi.org/10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cheung HW, Du J, Boehm J, He F, Weir B, Wang X, Butaney M, Sequist L, Luo B, Engelman J, Root D, Meyerson M, Golub T, Janne P, Hahn W. Amplification of CRKL Induces Transformation and Epidermal Growth Factor Receptor Inhibitor Resistance in Human Non–Small Cell Lung Cancers. Cancer Disc. 2011 doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- [43].Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. doi: 10.1126/science.1099314 1099314 [pii] [DOI] [PubMed] [Google Scholar]
- [44].Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Haura EB, Muller A, Brietwieser FP, Li J, Grebien F, Colinge J, Bennett KL. Using iTRAQ(R) Combined with Tandem Affinity Purification to Enhance Low-abundance Proteins Associated with Somatically-mutated EGFR Core Complexes in Lung Cancer. J Proteome Res. doi: 10.1021/pr100863f. doi: 10.1021/pr100863f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Massinen S, Tammimies K, Tapia-Paez I, Matsson H, Hokkanen ME, Soderberg O, Landegren U, Castren E, Gustafsson JA, Treuter E, et al. Functional interaction of DYX1C1 with estrogen receptors suggests involvement of hormonal pathways in dyslexia. Hum. Mol. Genet. 2009;18:2802–2812. doi: 10.1093/hmg/ddp215. doi: ddp215 [pii] 10.1093/hmg/ddp215. [DOI] [PubMed] [Google Scholar]
- [47].Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. doi: S1046-2023(08)00107-2 [pii] 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- [48].Melin J, Jarvius J, Larsson C, Soderberg O, Landegren U, Nilsson M. Ligation-based molecular tools for lab-on-a-chip devices. Nat. Biotechnol. 2008;25:42–48. doi: 10.1016/j.nbt.2008.02.003. doi: S1871-6784(08)00005-8 [pii] 10.1016/j.nbt.2008.02.003. [DOI] [PubMed] [Google Scholar]
- [49].Soderberg O, Leuchowius KJ, Kamali-Moghaddam M, Jarvius M, Gustafsdottir S, Schallmeiner E, Gullberg M, Jarvius J, Landegren U. Proximity ligation: a specific and versatile tool for the proteomic era. Genet. Eng. (NY) 2007;28:85–93. doi: 10.1007/978-0-387-34504-8_5. [DOI] [PubMed] [Google Scholar]
- [50].Jarvius M, Paulsson J, Weibrecht I, Leuchowius KJ, Andersson AC, Wahlby C, Gullberg M, Botling J, Sjoblom T, Markova B, et al. In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Mol. Cell. Proteomics. 2007;6:1500–1509. doi: 10.1074/mcp.M700166-MCP200. doi: M700166-MCP200 [pii] 10.1074/mcp.M700166-MCP200. [DOI] [PubMed] [Google Scholar]
- [51].Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. doi: nmeth947 [pii] 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- [52].Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. doi: nature06358 [pii] 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Meyerson M. Cancer: broken genes in solid tumours. Nature. 2007;448:545–546. doi: 10.1038/448545a. doi: 448545a[pii]10.1038/448545a. [DOI] [PubMed] [Google Scholar]
- [54].Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, Weinberg RA. Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 1999;5:1164–1170. doi: 10.1038/13495. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- [55].Li J, Lenferink AE, Deng Y, Collins C, Cui Q, Purisima EO, O’Connor-McCourt MD, Wang E. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat. Commun. 2010;1:34. doi: 10.1038/ncomms1033. doi: 10.1038/ncomms1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Broder S. Twenty-five years of translational medicine in antiretroviral therapy: promises to keep. Sci Transl Med. 2:39ps33. doi: 10.1126/scitranslmed.3000749. doi: 2/39/39ps33 [pii] 10.1126/scitranslmed.3000749. [DOI] [PubMed] [Google Scholar]
- [57].LoRusso PM, Anderson AB, Boerner SA, Averbuch SD. Making the investigational oncology pipeline more efficient and effective: are we headed in the right direction? Clin Cancer Res. 16:5956–5962. doi: 10.1158/1078-0432.CCR-10-1279. doi: 16/24/5956 [pii] 10.1158/1078-0432.CCR-10-1279. [DOI] [PubMed] [Google Scholar]
- [58].LoRusso PM, Schnipper LE, Stewart DJ, Boerner SA, Averbuch SD, Wolf W. Translating clinical trials into meaningful outcomes. Clin Cancer Res. 16:5951–5955. doi: 10.1158/1078-0432.CCR-10-2632. doi: 16/24/5951 [pii] 10.1158/1078-0432.CCR-10-2632. [DOI] [PubMed] [Google Scholar]
- [59].Koomen JM, Haura EB, Bepler G, Sutphen R, Remily-Wood ER, Benson K, Hussein M, Hazlehurst LA, Yeatman TJ, Hildreth LT, et al. Proteomic contributions to personalized cancer care. Mol. Cell. Proteomics. 2008;7:1780–1794. doi: 10.1074/mcp.R800002-MCP200. doi: R800002-MCP200 [pii] 10.1074/mcp.R800002-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Whitty A, Kumaravel G. Between a rock and a hard place? Nat. Chem. Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- [61].Fedorov O, Muller S, Knapp S. The (un)targeted cancer kinome. Nat. Chem. Biol. 2010;6:166–169. doi: 10.1038/nchembio.297. doi: 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- [62].Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr. Opin. Chem. Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. http://dx.doi.org/10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- [63].Walensky LD. From Mitochondrial Biology to Magic Bullet: Navitoclax Disarms BCL-2 in Chronic Lymphocytic Leukemia. J. Clin. Oncol. 2012;30:554–557. doi: 10.1200/JCO.2011.37.9339. doi: 10.1200/JCO.2011.37.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hwang JJ, Kuruvilla J, Mendelson D, Pishvaian MJ, Deeken JF, Siu LL, Berger MS, Viallet J, Marshall JL. Phase I dose finding studies of obatoclax (GX15-070), a small molecule pan-BCL-2 family antagonist, in patients with advanced solid tumors or lymphoma. Clin. Cancer Res. 2010;16:4038–4045. doi: 10.1158/1078-0432.CCR-10-0822. doi: 10.1158/1078-0432.CCR-10-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Roberts AW, Seymour JF, Brown JR, Wierda WG, Kipps TJ, Khaw SL, Carney DA, He SZ, Huang DC, Xiong H, et al. Substantial Susceptibility of Chronic Lymphocytic Leukemia to BCL2 Inhibition: Results of a Phase I Study of Navitoclax in Patients With Relapsed or Refractory Disease. J. Clin. Oncol. 2012;30:488–496. doi: 10.1200/JCO.2011.34.7898. doi: 10.1200/JCO.2011.34.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]