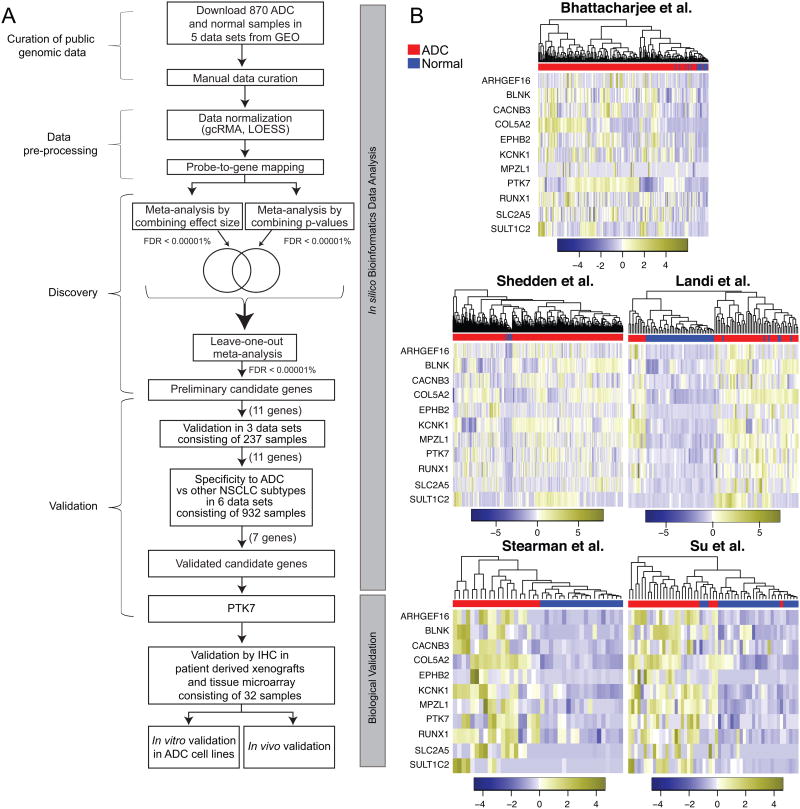
Figure 1.
Meta-analysis of NSCLC gene expression datasets identified 11 significantly over-expressed genes in NSCLC. A. Meta-analysis and functional validation pipeline. In silico bioinformatics data analysis workflow consisted of the curation of five publically available datasets, data pre-processing, discovery meta-analysis, independent validation, and subtype analysis followed by validation in primary human patient samples, and in vitro and in vivo functional validation. B. The 11 genes cluster ADC vs. normal across each independent dataset. Each column is a sample and each row is the expression level of a gene. The color scale represents the raw Z-score ranging from blue (low expression) to yellow (high expression). Dendrograms above each heatmap correspond to the Pearson correlation-based hierarchical clustering of samples by expression of the 11 genes.