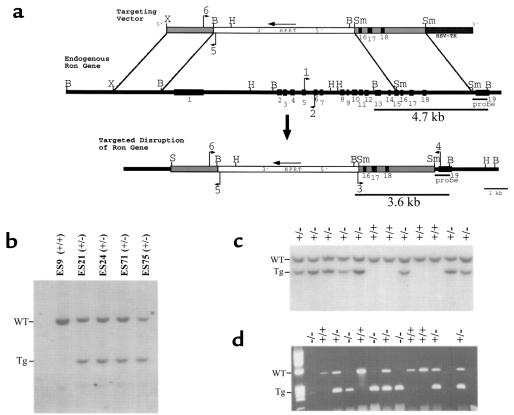
Figure 1.
Disruption of the mouse Ron gene in embryonic stem (ES) cells and mice. (a) Schematic representation and partial restriction map of the targeting vector (top) used to disrupt the endogenous mouse Ron gene (middle). The predicted organization of the targeted mouse Ron gene following homologous recombination is also shown (bottom). The targeting vector contains sequences homologous to the endogenous Ron gene (gray bars) flanking the HPRT expression cassette (white bar). The HSV-tk gene flanks the targeting vector at the 3′ end (dark gray bar). The exons of the endogenous Ron gene are numbered (1–19, black boxes). The primers used for PCR genotyping are indicated with arrows and are numbered 1–6. The probe used for Southern blot analysis is shown below the endogenous Ron gene. The BamHI fragments of genomic DNA that are complementary to the probe are indicated with the expected sizes (4.7 kb and 3.6 kb). B, BamHI. H, HindIII. S, SalI Sm, SmaI. X, XbaI. (b) Southern blot analysis of BamHI–digested DNA from selection-resistant ES cell clones. The wild-type allele (WT) is represented by a 4.7-kb fragment, while the targeted allele (Tg) is represented by a 3.6-kb fragment. ES9 was included as a wild-type control. (c) Southern blot analysis of BamHI–digested DNA from a single litter generated from a Ron+/– intercrossing. Offspring were genotyped at 7 days postnatal. Of the 12 littermates shown, 8 are Ron+/– (+/–) and 4 are Ron+/+ (+/+). (d) Genotype analysis by PCR of a single litter of E3.5 blastocysts generated from a Ron+/– intercrossing. The wild-type allele is represented by a 523-bp product generated from primers 1 and 2 (see a); the targeted allele is represented by a 274-bp product generated from primers 5 and 6.