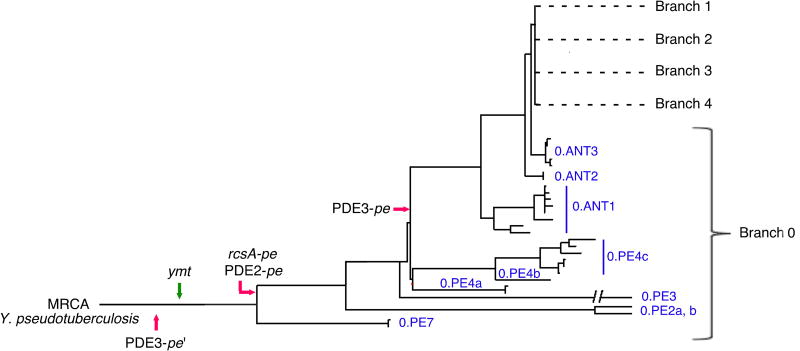
Figure 5. Early Occurrence and Fixation of Genetic Changes that Enabled Flea-borne Transmission During the Emergence of Y. pestis.
The earliest documented appearance in available genome sequences of gain of function (green arrow) and loss-of-function (red arrows) gene changes in the genealogy of Y. pestis is indicated. All changes are present in the Pestoides (0.PE) group in the root Branch 0 that are most closely related to the most recent common ancestor (MRCA), Y. pseudotuberculosis. The four changes in Y. pestis KIM are found in 0.ANT as well as all of the evolutionarily more recent Branch 1 and 2 strains. Genome sequences of the following were available for this analysis: Branch 0 strains 0.PE2a (Pestoides F), 0.PE3a (Angola), 0.PE4b (Pestoides A), 0.PE4c (91001), 0.ANT2a (B42003004) [The 0.PE7 strain contains ymt but the rcsA, PDE2, and PDE3 sequences were not accessible]; Branch 1 strains 1.ANTa (Antiqua), 1.ANT1b (UG05), 1.ANT1b (CP000308), 1.IN3g (E1979001), 1.ORI1 (A1122), 1.ORI3c (IP275), 1.ORI1d (CA88), 1.ORI1e (CO92), 1.ORI1g (FV-1), 1.ORI2u (F1991016), 1.ORI3b (MG05); Branch 2 strains 2.ANT1c (Nepal), 2.MED1a (KIM), 2.MED2c (K1973002), and 9 other unclassified biovar Orientalis strains. The figure and nomenclature are adapted from (Cui et al., 2013).