Abstract
Chromatin immunoprecipitation and deep sequencing (ChIP-SEQ) represents a powerful tool for identifying the genomic targets of transcription factors, chromatin remodeling factors, and histone modifications. The frogs Xenopus laevis and Xenopus tropicalis have historically been outstanding model systems for embryology and cell biology, with emerging utility as highly accessible embryos for genome-wide studies. Here we focus on the particular strengths and limitations of Xenopus cell biology and genomics as they apply to ChIP-SEQ, and outline a methodology for ChIP-SEQ in both species, providing detailed strategies for sample preparation, antibody selection, quality control, sequencing library preparation, and basic analysis.
Keywords: Chromatin immunoprecipitation, ChIP-SEQ, Xenopus tropicalis, Xenopus laevis, embryo
1. Introduction
At various points in its long history, the tractable Xenopus embryo has been at the forefront of embryology, reprogramming biology, cell biology, and cell cycle research, but is only now emerging as a genomic organism. Historically, the application of genome-wide sequencing approaches to Xenopus embryos, particularly to Xenopus laevis, has been hampered by the lack of a well-assembled, well-annotated genome. However, the current versions of the Xenopus tropicalis genome (1) and the rapid recent progress in assembly and annotation of the Xenopus laevis genome have made whole genome sequencing approaches quite feasible in both species, and there is now no practical limitation to applying these approaches in Xenopus.
Chromatin immunoprecipitation, first described in the 1980’s, has come to the fore as the preferred technique for analysis of transcription factor/DNA interactions and epigenetic modifications. In the last several years, chromatin immunoprecipitation combined with high-throughput sequencing (ChIP-SEQ) has yielded unprecedented amounts of data describing DNA/protein interactions in cultured cells and embryonic systems, most recently exemplified by the results of the ENCODE project in human cells, but also in yeast, Drosophila, cell culture systems, and, increasingly, in vertebrate embryology.
The unique embryology of Xenopus makes it an extremely powerful system for genome-wide analysis, particularly for questions of induction, fate specification, and dynamic processes. Explants of specific tissues allow investigation of transcription factor targets or epigenetic modifications with high precision in time and space. The ease with which hundreds or even thousands of stage-matched embryos can be obtained makes generation of samples from early embryonic stages or from specific tissue types easier in this system than other vertebrate embryos, allowing investigation of a broad range of transcription factors and developmental contexts. The extensive literature underlying Xenopus embryology, and the well-defined tools for studying patterning, morphogenesis and induction in early development, make the range of questions that could potentially be addressed with ChIP-SEQ in Xenopus essentially open-ended.
Xenopus also offers unique challenges to ChIP-SEQ, which we will discuss in detail. In young embryos, cells are very large and yolky, with extremely high protein:DNA and RNA:DNA ratios. Since the foundation of ChIP is isolation of clean nucleoprotein complexes, more effort must be dedicated to preparing Xenopus lysates for ChIP than for many other cell types. The paucity of available primary antibodies for Xenopus is a consideration as well, although there are several strategies for overcoming this limitation. In contrast to some model organism genomes, notably mouse, the assembly and annotation of the Xenopus genomes are poor, adding extra considerations when choosing programs for alignment, peak calling, and analysis. However, the Xenopus Genome Consortium is rapidly improving the state of both X. laevis and X. tropicalis genomes and we expect these issues to be transient (see http://www.xenbase.org/common/ for news updates and genome browsers). The combined efforts of researchers developing optimized protocols for ChIP and improved genomics for analysis make ChIP-SEQ in Xenopus currently practical, with the promise of rapid additional improvements in the near future.
In this methods overview we first outline a universal methodology for ChIP in both species, which uses features of several previously published protocols (2, 3) and highlights problems we have encountered, potential solutions, and troubleshooting approaches. We then describe a generalized method for ChIP-SEQ library preparation for the Illumina GA2 or HiSEQ platforms that works well for both Xenopus tropicalis and Xenopus laevis. Lastly, we suggest strategies for basic sequence analysis in both species – and as central to the entire methodology - provide insights into the current state and accessibility of both genomes. Detailed protocols follow each section.
1.2 Method overview and considerations before beginning
The wet lab portion of our ChIP-SEQ approach can be completed in approximately one week, although the time required to receive sequencing results and analyze them will vary and is dependent on the user’s needs. In brief, the steps are:
Chromatin preparation from embryos, which includes isolation of tissue, fixation/crosslinking, sample homogenization to remove yolk, and sonication. This takes 2 days. Note before beginning: we recommend optimizing sonication conditions before attempting ChIP with valuable samples or antibodies (see Section 2.4: Sonication).
Immunoprecipitation of crosslinked chromatin, including incubation of the sonicated embryo lysate with antibody-conjugated beads, washing, reversal of crosslinks, and DNA cleanup. These steps collectively take four days; the first two of which overlap with chromatin preparation. Note before beginning: choosing and validating antibodies that will perform well for ChIP takes some time, and in some cases generation of a tagged protein construct may be more feasible. See Section 3.2 Antibody choice, validation; antibodies versus tags, for detailed discussion of these considerations.
Preparation of the sequencing library, which includes end repair and adenylation of the ChIP pull down, adapter ligation, size selection, and amplification. Library preparation can be completed in one day, although there are several places where the DNA can be stored at −20°C. Note before beginning: the kits and resources for library preparation are improving rapidly, especially with regard to the starting amounts of DNA required for library preparation and purification protocols for size selection. We recommend users consult manufacturer websites and sequencing-related message boards (for example SEQanswers.com), for updated products and strategies.
Analysis. While we suggest a basic workflow and programs for analysis, our discussion will not replace the need for bioinformatics expertise. We note that Xenopus-specific bioinformatics training is offered each spring at the Xenopus Bioinformatics Workshop, given by the National Xenopus Resource at the Marine Biological Laboratory in Woods Hole, Massachusetts, which was quite successful in its inaugural session.
1.3 Controls
Later sections will discuss controls and validation methods for ChIP-SEQ, but some consideration of controls and quality control is useful at the outset of the experimental design. We recommend at minimum:
Quality control of the DNA, at least in initial experiments. Prior to library preparation, check input DNA for size, sonication completeness, and quality. This is discussed further in sections 2 and 4.
Validation of antibodies using Western blot. We have generally found that if an antibody cannot detect a clear target from embryo lysate on a Western blot, it will not work well for ChIP. Further antibody and tagging controls are discussed in section 3.
Plan experiments to include at least two biological replicates for each sample type (for example, unmanipulated and manipulated embryos, or embryos of differing stages, or explants versus whole embryos). This is useful at the stage of peak validation; ChIP-SEQ peaks that are present in both replicates and not in input libraries can be regarded as high-value.
Sequencing of input libraries. An “input” sample, representing chromatin that has not been immunoprecipitated, is collected after chromatin preparation for each sample (See section 2). ChIP-SEQ libraries made from these input samples will reveal the background distribution of chromatin fragments, and often show non-specific peaks that must be subtracted from ChIP analysis. In the analysis phase, the input library can be treated as the background level to compare with immunoprecipitation libraries. We have found that making a new input library for each set of experiments is essential. In the past, we have found that input libraries from embryos collected or sonicated on different days, even though apparently the same age or tissue type, can be different enough to conflate analysis. Pooling small batches of embryos from different collection times to create one sample and corresponding input library is fine, but if a completed experiment is repeated several weeks later a new input library should also be made. Another control that is often done is sequencing of libraries made with mock-pulldowns using IgG; with good input libraries we have not found this necessary.
Quantitative PCR validation of known transcription factor or histone target regions. If the ChIP target protein has known binding sites, we absolutely advocate confirming that these are enriched by ChIP-qPCR. However, we offer the caveat that the ChIP pull down DNA is precious, and we have frequently found that ChIP-SEQ reveals peaks in regions that did not show enrichment by qPCR. If the desired target region does not show enrichment, it may still be worth sequencing the sample; replicates, a good input library, and a well-chosen threshold for peak calling will reveal if peaks are specific.
2. Chromatin preparation
Our protocol combines features of previously published protocols for ChIP in Xenopus (2, 3). We have made some novel modifications to existing protocols that optimize them for sequencing - including issues concerning embryo numbers and sonication conditions.
2.1 Embryo collection
A range of embryonic starting material can be used for ChIP-SEQ in Xenopus. We have performed ChIP-SEQ throughout early development on whole embryos, explants, and MO-injected embryos. The numbers of embryos needed depends upon three factors – the number of cells, the abundance of the protein to be detected, and the quality of the antibody. We estimate that this protocol requires 3×105 −1×106 cells when examining an abundant protein with an excellent antibody (e.g., H3K4me3), and 1×106−1×107 cells when examining a transcription factor with a reasonably good antibody (e.g., Smad2/3; (4)). While these numbers seem daunting, consider that a stage 8 embryo contains approximately 5,000 cells(5–7) requiring the harvesting of between 60 and 2,000 embryos depending upon the target protein’s abundance and antibody quality, which is not a difficult task. We also find that for excellent antibodies, 300 animal caps are sufficient for ChIP. A consideration with explants is that while 300 animal caps isolated at stage 9 represent approximately 1×105 cells, this number expands rapidly as the explants age, such that 300 animal caps harvested at stage 10.5 represent approximately 1×106 cells. As a generous starting point for ChIP-SEQ using endogenous transcription factors, we recommend using 1000 X. tropicalis embryos stage 9 or older, 500 X. laevis embryos stage 9 or older, and 600 animal cap explants. Empirically, we have found that successful ChIP requires fewer Xenopus laevis embryos than Xenopus tropicalis embryos of the same stage. This may simply be due to the larger genome size of X. laevis providing more total chromatin, but the total cell or protein volume may also be a factor. For histone modifications or tagged proteins, these numbers can be halved, while for embryos younger than stage 9, or for antibodies that perform poorly in Western blots, they should be doubled. These quantities should reliably give yields of at least 1µg of DNA after chromatin immunoprecipitation, which is reliably sufficient for production of very good libraries.
2.2 Crosslinking
The first critical step in ChIP is to crosslink the protein to the DNA. We have reliably followed the crosslinking protocol established by others (3), which we highlight here with a few comments. Embryos are treated for 1 hour in a solution of 1% formaldehyde diluted in 1× PBS. We use freshly-made fixation solution, with high quality methanol-free formaldehyde. As in other protocols, we find that the fixation time is an important factor in the size and polydispersity of the chromatin fragments recovered, as well as the success of the immunoprecipitation. Xenopus cells require longer fixation times to achieve significant crosslinking than do cultured cells or yeast (1,2). While 45 minutes or 1 hour and 15 minutes still result in high-quality ChIPs, we don’t recommend shorter or longer fixation times. The number of embryos that can be fixed at one time is flexible—we have had comparable success fixing batches of 20 or 1000 embryos, typically in 1 dram glass vials. Following fixation, the reaction is quenched by a washing in 0.125M glycine/PBS for 5 minutes, followed by 3 quick washes in PBS. All liquid is then removed from the embryos, and they may be either processed immediately for ChIP, or frozen, dry, for up to 6 months at −80 or in liquid nitrogen.
2.3 Sample homogenization and yolk removal
After crosslinking, embryos or explants must be homogenized in order to make the nuclei accessible to sonication. Crosslinked embryos (thawed if previously frozen) are homogenized thoroughly in cold RIPA buffer with added protease inhibitors by pipetting up and down with a P1000 tip or with a plastic pestle. We typically process embryos in batches with up to 100 Xenopus laevis or 250 Xenopus tropicalis stage 10.5 embryos per tube. During homogenization, avoid generating bubbles by keeping the pipette tip beneath the surface. Exposure of proteins to the air interface can denature them, even if the bubbles are subsequently removed (8).
For Xenopus, in particular, homogenization is important for removing the bulk of yolk while leaving nucleoprotein complexes intact. Because Xenopus tissue is yolky and the cytoplasm to DNA ratio is so high, more preparation is required for Xenopus prior to sonication than is required for other ChIP protcols. We find complete removal of the yolk to be a persistent problem, but several modifications have made significant improvements. Two earlier recommendations from (3) are central to the adaptation of the ChIP protocol to Xenopus: first, repeat homogenization, followed by centrifugation, helps remove yolk protein. To remove the yolk from the sides of the microfuge tube, wrap a plastic pestle in a kimwipe, and carefully wipe the inside of the tube after decanting the supernatant, taking great care not to touch the pellet. Second, lowering the SDS level in RIPA buffer to 0.1%, as opposed to 1% in standard protocols, greatly improves yield.
2.4 Sonication
The sonication conditions are critical for the generation of a successful ChIP, and must be determined empirically. Relative to cultured cells – or even mammalian tissues – Xenopus embryos are highly resistant to sonication. Optimizing our own sonication conditions took considerable time, and each new user has made small adjustments to suit their own results. Like others, we have found substantial variation in sonication effectiveness between sonicator types, between individual sonicators of the same type, and even between users of the same sonicator. While we describe our sonication conditions here, the specific conditions used for each sonicator will be different, and should be chosen by analysis of the resulting DNA fragments. When attempting ChIP for the first time, we recommend that users try out a range of crosslinking and sonication conditions on wild-type embryos in a dry-run, without attempting to ChIP the DNA, to optimize the conditions without wasting valuable injected embryos, antibody, or time. This simply means fixing embryos for a range of times (for example 30, 45 and 60 minutes), clearing the samples, sonicating for a range of cycle numbers (for example 2,5, and 10 cycles at 30%, 50% and 100% intensity), then reversing crosslinks, precipitating the DNA, and analyzing the resulting DNA electrophoretically for yield and degree of shearing. Appropriately sonicated DNA should run as an easily-visible smear on a 2% agarose gel, with a mean fragment size of 300–500bp. Excellent visual examples of well- and poorly-crosslinked and –sonicated DNA are available in earlier protocols (3).
A persistent problem we have confronted with sonication is the sample foaming—this can be caused by the tip coming too near the surface or by sample overheating. Both foaming and overheating are damaging to nucleoprotein complexes and should be avoided. We have found that pre-chilling the samples thoroughly helps considerably. A combination of ice and water (wet ice) works far better than ice alone. Lysates should be chilled on wet ice for at least 20 minutes before sonicating. Immediately before sonicating, they should be resuspended by gently pipetting up and down. If using a tip sonicator, the sonication can be done directly on wet ice, with tubes secured by holding the cap while the tube is loosely secured in a round plastic tube rack. To provide an example of sonication conditions (remember they will be different for each user), we use a Branson 450 sonifier tip sonicator, and perform 5 rounds of sonication on each tube of embryo extract (up to 100 stage 10.5 embryos for X. laevis or 250 for X. tropicalis), with each round consisting of 20 seconds at 100% intensity. Notably, previous reports using a similar sonicator tip required fewer rounds and intensity of sonication (3), highlighting the user specificity of sonication conditions. Between rounds of sonication, each sample must be re-chilled on wet ice for at least 1 minute. While bath sonicators might also be used, we have found that many more rounds of sonication (up to 20 or more) are necessary to achieve the same size range of chromatin fragments, and have had better success with a tip sonicator on full intensity.
Following sonication, the lysate samples are centrifuged to pellet all cellular debris. Some of the supernatant is reserved for input controls. These will be used for control library preparation, but can also provide valuable information on overall yield and sample quality of the sonicated DNA. Sonicated DNA should run as an easily-visible smear with a mean size of 300–500bp.
2.5 Chromatin preparation materials and equipment
Methanol-free formaldehyde (Sigma F8775)
0.125M Glycine
1× PBS
Magnetic particle concentrator (Invitrogen 123.21D)
Dynabeads protein A (Invitrogen 10001D)
Dynabeads protein G (Invitrogen 10003D)
Antibody (diluted according to manufacturer instructions)
PBS+5% BSA: 250mg BSA (Sigma A9647-50G) in 50ml PBS (Gibco 10010-023). Store at 4° and discard after one week.
RIPA buffer (4°C, 1.25 ml per set of 50 embryos): 50 mM Tris-HCl, pH 7.4, 1% Igepal CA-630 (NP-40) (Sigma I3021), 0.25% Na-Deoxycholate, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.5 mM DTT, 5 mM Na-Butyrate
Complete, Mini Protease Inhibitor Cocktail Tablets (Roche 11836153001), 1 tablet per 10mL RIPA buffer.
Rotator or Nutator, at 4°C
Sonicator
Refrigerated centrifuge
2.6 Detailed chromatin preparation procedure
Day 1:
Culture embryos to desired stage.
Fix 1 hour in 1% formaldehyde/PBS
Wash 5 minutes in 0.125M glycine
Wash 3 times, quickly, in PBS. Divide embryos into batches of up to 100 X. laevis or 250 X. tropicalis.
Remove all PBS and freeze up to 6 months at −80°C, or proceed directly to Day 2 steps.
Prepare Dynabeads with antibody (see section 3)
Day 2:
Thaw crosslinked embryos on ice, 10–15 minutes.
Add 600µl cold RIPA+Protease Inhibitor to each sample.
Break embryos by pipetting with a P1000 tip or by gentle disruption with a plastic pestle, until embryos are broken into small fragments and the solution is gray in color. The embryos need not be completely homogenized yet.
Centrifuge at 14,000×G for 10 minutes using a refrigerated centrifuge.
Decant supernatant, and wipe the walls of the tube with a kimwipe to remove traces of yolk.
Add 100µl cold RIPA, and homogenize thoroughly, taking care to avoid bubbles, until no visible fragments of embryo remain and homogenate is a uniform gray.
Add 550µl cold RIPA.
Chill embryo samples on wet ice for 20–30 minutes, until ready to sonicate. Gently resuspend each sample by pipetting immediately before sonication.
Sonicate using empirically determined conditions. We recommend trying 2, 5, or 10 cycles at 30%, 50% or 100% intensity to start.
Centrifuge at 14,000×G for 10 minutes using a refrigerated centrifuge.
Transfer 60µl of the resulting lysate to new tubes for input controls.
Transfer supernatant to pre-clear beads, and then to antibody-conjugated beads (see section 3).
3. Immunoprecipitation
3.1 Antibody-conjugated bead preparation
The first step in immunoprecipitation is conjugation of the primary antibody to a substrate that can easily be precipitated. While there are several reagents available for this purpose, we recommend magnetic Dynabeads, which give high yields and are simple to use. We use protein G-coupled Dynabeads for histone modifications, and protein A-coupled Dynabeads for transcription factors. For detailed information on which Dynabead product to use, see http://www.invitrogen.com/site/us/en/home/Products-and-Services/Applications/Protein-Expression-and-Analysis/Protein-Sample-Preparation-and-Protein-Purification/ProteinSPProteinIso-Misc/Protein-Isolation/Immunoprecipitation-FAQs.html
3.2 Antibody choice, validation; antibodies versus tags
The antibody is hands-down the most important aspect of a successful ChIP. Without a functioning antibody, ChIP will not work and much time and money will be wasted trying to validate ambiguous results. Clearly, tried and true antibodies that have been previously used to perform Western blotting and immunocytochemistry in frogs are the best reagents to use. However, there are few validated Xenopus antibodies, so many of us will have to make our own antibodies or run the gauntlet by purchasing from companies. We’ve had surprising success using the later strategy, but have had to purchase multiple antibodies – for both transcription factors and histone modifications – to find one that actually works. Clearly antibodies advertised as Xenopus validated and ChIP-validated antibodies are the most desirable, although we have had success purchasing non-Xenopus ChIP-validated antibodies where the target epitope is conserved in Xenopus. Certainly it is critical to test any antibody by Western blot prior to performing the first ChIP, and confirm that a clear target band of the expected size is present in embryo lysate. For additional rigor, or with newly-created antibodies, it’s ideal to check that the antibody target is lost in knockdown conditions. While there may be antibodies that work for ChIP, but not Western, our opinion is that this is rare, and if the antibody doesn’t work to detect protein on a Western, it is unlikely that it will work for ChIP.
Once the antibodies are chosen and the ChIP performed, the question becomes whether the DNA pulled down is specific. While we do use the standard validation approach of qPCR for known direct targets of the transcription factor, we have frequently found (in Xenopus, mouse and human cells) that ChIP-PCRs often show little enrichment, even though sequencing of that sample will reveal a peak at the PCR target site. Therefore, one is left with the conundrum of whether to proceed to sequencing if the qPCR provides a negative result. With the increasingly manageable cost of high-throughput sequencing and the preciousness of ChIP DNA, we find that sequencing a valuable ChIP sample may be preferable to extensive qPCR validation.
If an antibody is simply not available, then overexpressing an endogenous level of a tagged proteins is an excellent option. Many ChIP validated tags exist for Xenopus, including eGFP, myc, and FLAG. With tagged proteins, additional controls to confirm the function, dose and specificity of the fusion protein are necessary. Our collaborators at UC Berkeley have recently had success performing ChIP with β-catenin using a C-terminal FLAG tag (J. Young, pers. communication). As general controls for fusion proteins, they recommend checking the fusion protein by Western blot to confirm the tag is detectable and is in frame, then confirm functionality and specificity of the tagged fusion by using it to recapitulate overexpression phenotypes and to rescue morpholino knockdown of the target. These experiments will also indicate what dose of fusion protein mRNA would be tolerated well by the embryo and would be comparable to endogenous protein levels, although we note that recent evidence in yeast suggests overexpression of transcription factors does not lead to excessive false positives in ChIP (9).
3.3 Lysate incubation
To avoid any non-specific binding of chromatin fragments to Dynabeads, sonicated, centrifuged embryo lysate is first incubated with control Dynabeads, to “pre-clear” the lysate. The lysate is then transferred to antibody-conjugated Dynabeads, and incubated at 4°C overnight.
3.4 Clean-up, reversing crosslinks, DNA recovery and quality control
After the antibody has had time to bind to the chromatin, the chromatin fragments and DNA are handled similarly to any other ChIP protocol, as the specific foibles of Xenopus cell biology are no longer in play. Following binding of chromatin fragments in the embryo lysate to antibody-conjugated beads, nonspecific chromatin interactions are removed with several wash steps of increasing stringency. After washing, chromatin is released from the beads with vigorous vortexing in an SDS-containing buffer. To make sure the chromatin fragments are fully recovered, we perform several rounds of vortexing at room temperature and at 65°C. At this point, the Dynabeads are discarded, and the DNA is incubated overnight in high salt solution at 65°C to reverse crosslinks. Blythe et al. perform the overnight incubation with proteinase K and glycogen. For additional DNA cleanup, of DNA that will subsequently be used for qPCR, we recommend including proteinase K and glycogen in this overnight incubation (3).
3.5 DNA purification and quality control, qPCR notes
Following reversal of crosslinks, the DNA can be purified and isolated in several ways. The choice of method depends somewhat on the experimental design and the reactions that will be done with it. We have found that for experiments with large numbers of embryos, where the DNA will be used directly for sequencing, phenol/chloroform extraction and ethanol precipitation is sufficient. However, if the experiment uses very young embryos, where the Protein:DNA and RNA:DNA ratios are very high, or in experiments where some analysis will be done by qPCR, it is preferable to have more rigorous purification standards, including proteinase K and RNAse treatments and an additional purification step over a Qiagen Minelute column.
3.6 Immunoprecipitation materials
Low salt buffer: 0.1% SDS, 1% TritonX-100, 2mM EDTA, 20mM Tris-HCL pH8.0, 150mM NaCl (Store at 4°C)
High salt buffer: 0.1% SDS, 1% TritonX-100, 2mM EDTA, 20mM Tris-HCL pH8.0, 500mM NaCl (Store at 4°C)
LiCl salt buffer: 0.25M LiCl (L-8895 Sigma), 1% IGEPAL CA630 (Sigma I-3021), 1% deoxicholate acid, 1mM EDTA, 10mM Tris-HCL pH8.0 (Store at 4°C)
TE buffer: 10mM Tris-HCl, 1mM EDTA, pH 8.0 (Store at 4°C)
TES buffer: 50mM Tris-HCl pH8.0, 10mM EDTA, 1% SDS (store at room temperature)
5M NaCl
Phenol/Chloroform/Isoamyl Alcohol (Thermo Fisher BP1752)
Chloroform (Thermo Fisher BP1145)
Sodium Acetate buffer solution (Sigma S-7899)
Glycogen (Fermentas cat# R0561 Fermentas)
Qiagen PCR Purification Kit (Qiagen 28004)
RNase A (Roche 10109169001)
70% and 100% Ethanol
3.7 Detailed immunoprecipitation procedure
Day 1 (the day before homogenization and sonication):
Resuspend protein G or protein A Dynabeads by vortexing. Use 50µl dynabeads for each experiment, including mock pulldown, if performing. All dynabeads can be aliquoted in one volume and washed together.
Place tubes in bead separator, wait until beads have migrated against magnet and sample is clear.
Wash beads twice with 1ml of PBS+5% BSA, vortexing beads well and then returning to magnet for each wash.
Resuspend beads in PBS+5% BSA, using 200µl for each experiment. Vortex beads well and aliquot into individual tubes for each experiment. Bring volume up to 1ml (+800µl) with PBS+5% BSA.
Add primary antibody according to manufacturer-recommended dilutions.
Place tubes on a nutator or rotator at 4°C overnight.
Day 2 (same day as sonication):
Prepare 20µl of washed Dynabeads per sample as above, resuspending in 1 mL PBS+5% BSA. These are the “pre-clear” beads
Place against magnet to pellet pre-clear beads, discard supernatant, and replace with the supernatant of sonicated, centrifuged embryo lysate.
Incubate pre-clear beads and lysate with rotation at 4°C for one hour.
Use magnet to pellet ANTIBODY-conjugated beads. Remove and discard supernatant. Use magnet to pellet PRE-CLEAR beads, and transfer supernatant (lysate) from pre-clear beads onto antibody- conjugated beads. Discard pre-clear beads.
Incubate antibody- conjugated beads plus lysate 4°C overnight, with rotation.
Day 3:
Use magnet to pellet antibody-conjugated beads, discard supernatant. Perform washes at 4°C, using 1 ml of solution for each wash, and using the magnet to pellet beads between each wash:
Low salt solution (2 washes, 5 minutes each)
High salt solution (2 washes, 5 minutes each)
LiCl buffer (2 washes, 5 minutes each)
TE (2 washes, 5 minutes each)
Remove all TE, and replace with 200µl TES. Return to room temperature.
Vortex beads very thoroughly, let settle, and vortex again.
Incubate beads at 65 degrees for 15 minutes, vortexing every 5 minutes.
Vortex thoroughly once more, pellet beads using magnet, and transfer supernatant carefully to new, labeled microfuge tube.
Remove input samples from freezer and thaw.
-
Add 16µl 5M NaCl to input samples and immunoprecipitated samples. Cap lids tightly and incubate at least 5 hours or overnight at 65°C.
*Optional: for higher-quality DNA, include 7µl proteinase K/Glycogen solution.
Day 4:
Add 1 volume (approx. 200µl) Phenol/Chloroform to samples. Vortex until milky, and centrifuge at 12,000×G for 5–7 minutes.
Transfer supernatant to new, clean, labeled microfuge tube.
Repeat extraction with 1 volume (200µl) Chloroform; vortex, spin and transfer supernatant as above.
Add 1/10 vol (20µl) 3M Sodium acetate, 2.5 vol (500µl) 100% Ethanol, and 1µl glycogen. Precipitate at least 5 hours or overnight at −20°C.
Centrifuge at full speed for 15 minutes, taking care to note the orientation of the tubes. Small pellets should be visible in input samples, but may not be visible in IP samples.
Carefully remove supernatant and wash pellets with 500µl 70% ethanol.
Centrifuge at full speed for 1 minute, taking care to note the orientation of the tubes and position of pellets. Carefully remove all traces of supernatant.
Resuspend in 15µl of nuclease-free water and quantify yield as described below. Alternatively, if the resulting DNA will be used for qPCR, perform the following additional steps:
Incubate in 100ul RNAse A/TE for 1 hour at 37°C.
Purify using a Qiagen Minelute reaction purification kit, according to kit instructions, using 15µl as the final elution volume
Quantify yield using nanodrop. If yield is low (<50ng/µl), use a high-sensitivity method such as Qbit to accurately quantitate yield.
4. ChIP-SEQ library preparation
There are several potential platforms for ultra high-throughput sequencing of ChIP-SEQ libraries. We prefer paired-end libraries for use on an Illumina GAII or HiSEQ high-throughput sequencing platform (see “6.1 Sequencing Platform Considerations”, below). We use Illumina’s “Genomic DNA Sample Prep Kit,” with some modifications. The recently released TruSEQ kit from Illumina is very promising for multiplexed/indexed/”barcoded” samples, but we haven’t yet worked with it enough to offer a detailed recommendation. For labs preparing many high-throughput sequencing libraries of multiple types, it may be more cost-effective to purchase the enzymes and reagents individually rather than as a kit. We also note that the Illumina kit supplies the bare minimum of reagents (including some inexpensive reagents like buffers) for 10 libraries. Therefore, while the Illumina kit offers the advantage of high quality control and reproducibility, we have also included purchasing information for individual reagents.
4.1 Starting DNA quantity and quality
The quality of the starting DNA must be reasonably high, but need not be excessively so. Illumina recommends that the starting OD 260/280 ratio should be approximately 1.8, but we have successfully made libraries from samples with ratios of 1.5 when concentrations are low. While we recommend beginning with high-quality DNA when possible, we find that for low-yielding ChIPs (due to small amounts of material or poor antibodies), it is better to use a larger quantity of poor-quality DNA than further deplete yield with additional purification steps.
The kit is optimized for starting quantities of 1µg–5µg total DNA. We have found it is possible to prepare libraries with 0.5µg of starting DNA using this kit, but this library required 16 cycles of PCR amplification, which noticeably reduced library complexity. The lowest amount of starting DNA we have attempted to make a library with is 0.3µg, and in this case the resulting library was unusable, with low complexity resulting from overamplification of few genomic regions. The current generation of TruSEQ kits available from Illumina promise libraries from initial DNA amounts of as little as 10ng; even if this low a target proves unrealistic, we expect it should soon be routine to prepare libraries from 50–100ng of DNA.
4.2 End repair, poly A-tailing, adapter ligation
While the kit begins with a genomic DNA shearing step, this is dispensable for sonicated DNA fragments, which are already less than 1kb. We begin with end repair, followed by A-tailing and adapter ligation. The amount of adapter added to the reaction should be stoichiometrically matched to the amount of starting DNA, although we have found that this is fairly forgiving. These steps require purification of the reaction between steps, so that each enzyme can work in compatible conditions. Illumina recommends the use of Qiagen Minelute columns for this purpose, which we use. In contrast to standard Qiagen prep columns, the Minelute column can efficiently elute DNA in as little as 8ul of volume.
4.3 Gel purification
After adapter ligation, excess adapters must be removed from the reaction, and the library should be size-selected. Both steps are accomplished at once by gel-purification of the adapter-ligated library. Illumina favors an insert size of 200+/− 25bp, and we follow this recommendation. The sample is run out on a gel, and the portion of the sample between 200–250bp is isolated from the gel. The DNA should be visible, sometimes faintly, as a smear. For accurate sizes-election, a 50bp or 25bp ladder is preferable to a 100bp ladder.
We have generally followed Illumina’s recommendation of a 3% agarose gel purification, but have also experimented with the e-Gel purification system, using 2% e-Gels. The e-Gel system is much faster, as the resulting DNA can be used directly for PCR and does not have to be extracted from the agarose. A problem we have encountered with e-Gels is that the range of the region excised is typically very small; only a fraction of the total 200–250bp range. To combat this, one can excise several bands of similar size (adding 20ul of additional water to the collection well after each band isolation), or run the e-Gel in reverse to capture the same region a second time; both of these result in a dilute sample that may need to be further concentrated by running over a Minelute column. In general, our recommendation is that if the starting amount of ChIP DNA was high (>2µg), the time efficiency of the e-Gel is worth the reduced yield, while if the starting amount of DNA was less, standard agarose gel-purification using a Qiagen gel purification kit is preferable. An issue to be aware of with either type of gel purification is that libraries can be cross-contaminated if run on the same gel. This is easily avoided by leaving at least one empty lane between each sample, and between samples and the ladder.
4.4 PCR amplification
After gel purification, the size-selected library is amplified with between 10 and 16 rounds of PCR, using Illumina’s primers. For indexed/”barcoded’ samples, a range of reverse primers are available, which Illumina will supply upon request. We recommend consulting information available through the Illumina website in order to optimize reverse primers, as the choice of primers varies with the number of samples barcoded, and must be balanced for the two laser colors of the sequencer.
Illumina’s kit recommends 10 cycles of amplification, but we don’t usually find this to yield enough for a good library. We have found that 1µg of starting DNA makes a fine library with 13–15 cycles of amplification, and that with a starting amount of 2–3µg of DNA, 12 cycles is sufficient. (While, in theory, 1µg of starting DNA should require one extra cycle relative to 2µg, and 0.5µg should require one cycle more than 1µg, we find that the amplification yields don’t scale in this tidy fashion, perhaps due to differential losses on columns). The PCR amplification begins with only 4µl of size-selected DNA, although the size selection step yields 20–30µl. Therefore, one way to improve yield without increasing the number of cycles is to perform several PCR reactions and pool them, which we have found to be successful.
The inevitable tradeoff with library preparation is quantity of starting material versus the amount of amplification. Less amplification requires a higher amount of starting material, and more amplification results in reduced library complexity. We have favored better library complexity, and therefore start with high amounts of starting material with less amplification. The other constraint that must be considered is the requirements of your sequencing facility. Our sequencing facility will not accept final library amounts less than 30ng (10µl of 3ng/µl sample). If your requirements are more permissive, library preparation from smaller starting quantities may be more realistic.
After PCR, the remaining primers should be removed from the library, or they will represent a high fraction of the sequenced sample. This can be achieved with a second round of gel purification, but purification over a Minelute column also works well, leaving only small amounts of unincorporated primers.
4.4 Library quality control
In quantitating the yield of the library, we have found that nanodrop is not accurate. While part of the reason is that OD ratio-based calculations of yield cannot discriminate between library DNA and residual unincorporated primers or adapters, the more significant reason is that nanodrop simply overcalculates yield at low concentrations (<50ng/µl), in some cases by as much as 10 fold. Our library yields, when calculated accurately, are typically 3–20ng/µl. A more sensitive calculation method uses the high-sensitivity double-stranded DNA application of Qbit, but the best method for calculation is Bioanalyzer, which gives more metrics about library quality, including concentration, mean fragment size (should be slightly larger than the size isolated in gel purification; about 250bp), and a sense of the amount of primer/adapter contamination.
4.5 Library preparation materials
All inclusive kits:
Genomic DNA prep kit from Illumina (requires Minelute reaction cleanup reagents and columns as well, below)
TruSEQ kit from Illumina
Piecemeal:
T4 ligase and buffer. Invitrogen 15224-017
T4 DNA Polymerase. NEB M0203L
10mM dNTPs. Invitrogen 18427-013
Klenow DNA Polymerase
T4 PNK
Minelute reaction cleanup kit. Qiagen 28204
dATP. Invitrogen 10216-018
Klenow Fragment (3’–5’ Exo). NEB M0212L
HPLC-purified Adapters (consult Illumina for sequences)
E. coli DNA ligase. Invitrogen 18052-019
2% eGel. Invitrogen G6610-02 –or-
NuSieve GTG agarose. Lonza 50080
50bp ladder. Invitrogen 10416-014
Phusion high-fidelity 2× master mix polymerase kit. NEB M0532S
HPLC-purified primers (consult Illumina for sequences)
4.6 Detailed Library prep procedure
Day 1:
Aliquot enough ChIP DNA to reach 1µg (minimum) or 5µg (ideal). Bring up DNA samples to 30ul with nuclease-free water.
- Repair ends by preparing the following reaction mix, using PCR tubes:
ChIP DNA 30µl Nuclease free water 45µl T4 ligase buffer 10µl dNTPs 4µl T4 DNA polymerase 5µl Klenow DNA Polymerase 1µl T4 PNK 5µl Total 70µl Incubate at 30°C for 20 minutes
- Clean up the reaction using Qiagen Minelute reaction clean up kit, as follows (or follow manufacturer instructions):
-
-To the 70µl reaction mix, add 300µl buffer ERC, mix.
-
-Pipet into Minelute column inside collection tube.
-
-Centrifuge at full speed one minute.
-
-Discard flow-through.
-
-Add 750ul Buffer PE to the top of column, centrifuge 1 minute.
-
-Discard flow through, and centrifuge again 1 minute to remove traces of ethanol.
-
-Transfer Minelute column to new, clean, labeled microfuge tube.
-
-Add 32µl buffer nuclease-free water to the middle of each column, let sit one minute.
-
-Centrifuge 1 minute at full speed.
-
-
Transfer samples to PCR tubes.
- To each sample (32µl), add:
Klenow Buffer 5µl dATP 10µl Klenow exonuclease (3’ to 5’ exo) 3µl Total 50µl Incubate at 37°C 30’.
Clean up reaction using Minelute column as above, eluting in 18µl final volume.
Transfer samples to PCR tubes.
- Ligate adapters by preparing the following reaction mix:
End-repaired DNA 18µl DNA ligase buffer 25µl Adapter oligo mix 2µl DNA ligase 5µl Total: 50µl React at 15°C for 20 minutes.
-
Gel-purify samples using either a 2% e-Gel or 3% Nuseive Agarose gel (see above).
For eGel:-
-Load samples into upper wells, leaving a space between each sample to avoid contamination. Choose one lane to run a 50bp ladder.
-
-Load 20ul water into any empty top wells, and all bottom wells.
-
-Select “2%” mode. Run eGel for 13 minutes.
-
-Use UV illumination to check position of ladder. DNA in samples should be visible as a smear. Add 10µl water to each bottom well.
-
-Run gel slowly, checking frequently, until bottom wells are positioned between the 200 and 250 bp band. Isolate these bands from each sample. If the gel runs too long, change mode to “reverse e-Gel” and run in reverse until the desired band comes into the bottom wells.
-
-If desired, add 20µl additional water to bottom wells, and run gel until 300bp band is positioned in bottom well. Isolate these bands from samples.
-
-
- For Agarose:
-
-Prepare 3% gel using NuSeive GTG agarose and TAE
-
-Combine samples with 10× loading dye. Also prepare a 1:10 dilution of 50bp ladder with loading dye.
-
-Load samples, leaving an empty well between each sample to avoid contamination.
-
-Run gel according to usual methods. Check by UV visualization; the DNA should be visible as a smear in wells containing samples.
-
-Use a clean scalpel blade to isolate the 200–250bp region.
-
-Purify DNA from gel fragment using Minelute gel purification kit, following manufacturer instructions, and eluting in 20µl final volume.
-
-
- Amplify the library by preparing the following PCR reaction:
Gel-purified DNA 4µl Phusion DNA polymerase 2× Master Mix 25µl Primer 1.1 1µl Primer 2.1 1µl Water 19µl Total 50µl - Perform the following PCR reaction, using 15 cycles if the initial amount of ChIP DNA was 1µg, or 12 cycles if the initial amount of DNA was 2µg or more.
- 30 seconds at 98°C
- [10 seconds at 98°C
- 30 seconds at 68°C
- 30 seconds at 72°C] × 12–15 cycles
- 5 minutes at 72°C
Purify the resulting PCR products using a Minelute PCR cleanup kit, following manufacturer instructions, eluting in 20µl final volume.
Check DNA concentration using a high-sensitivity method, such as Qbit or Bioanalyzer. If concentrations are too low (below 3ng/µl), repeat PCR and combine and concentrate reactions.
5. State of the genomes
As of this writing, genome browsers are available from the Xenbase homepage (http://www.xenbase.org/common/) for both Xenopus species, and include browsers for both the 2005 release (v4.1/Xentro2) and 2009 release (v7.1/Xentro3) for Xenopus tropicalis as well as the recent draft 2012 release (v6.0) of Xenopus laevis. Custom tracks can be uploaded directly to these browsers, which also include the UCSC browser for older version of the X. tropicalis genome (4.1/Xentro2). The draft version of the Xenopus laevis genome (6.0) is also available to download from Xenbase, (ftp://ftp.xenbase.org/pub/Genomics/JGI/Xenla6.0/) while the X. tropicalis genome is accessible through NCBI and UCSC (http://hgdownload.soe.ucsc.edu/downloads.html#xentro). The Xenopus laevis genome is under eager and rapid development, with at least one new release (v.7) likely before this article reaches print (T. Kwon and A. Session, pers. communication).
A few aspects of the state of the genome assembly can impact the ease and success of ChIP-SEQ analysis, and so we have therefore chosen to present some of the relevant genome statistics that may directly impact ChIP-SEQ in Xenopus. We outline several possible scenarios where poor genome assembly can impact analysis (Figure 1).
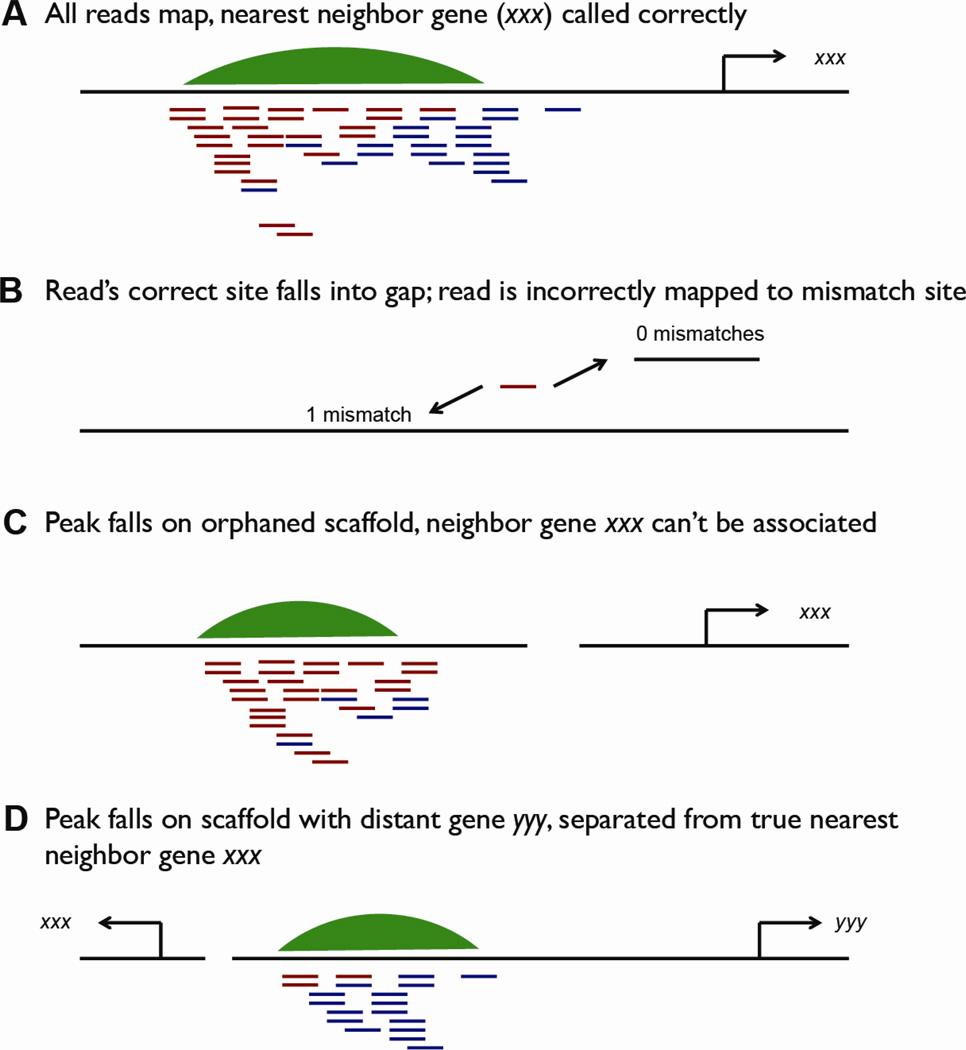
Figure 1.
Potential scenarios that may interfere with sequencing analysis. With an incompletely assembled genome, several categories of errors may occur in attempting to map reads and assign peaks to neighboring genes. A) Best-case scenario. Sequencing reads (red and blue lines indicating + and − orientations) are uniquely mapped to their positions in the genome (black bar), generating a smooth pileup that is called as a peak (green). That peak is putatively assigned to its nearest neighboring gene, gsc in this example. B) If the best match for a read (red line) falls into a gap (small black bar), usually those reads will simply fail to map, but the read may be erroneously mapped to a different region, especially if single mismatches are allowed in the alignment algorithm. C) If a peak falls on a small scaffold, it may be orphaned from the gene it corresponds to, and information about regulatory regions for that gene will be lost. D) Alternatively, if a peak is flanked by two genes, but a scaffold break falls between the nearest target and the peak, that peak may be inappropriately assigned to the other flanking gene. This scenario is mitigated somewhat by the fact that not all actual target genes are the closest neighboring genes to potential regulatory regions.
Ideally, sequencing reads would be mapped to their best possible match in the genome, resulting in a pileup of reads that can easily be called as a peak, and associated to a nearby, annotated gene (Fig 1A). Therefore, the first place that analysis can go awry is at mapping—if the best match for a read is in a gap in the genome assembly, the read will either not be mapped, or, depending on the mismatches allowed by the mapping algorithm one chooses, may be mapped in error to a position with a mismatch (Fig 1B). This is currently a moderate but not severe problem in Xenopus laevis: the current version 6 has 10.8% gaps, while the upcoming version 7 is anticipated to contain 10.1% gaps (A. Session, pers. communication). This suggests that approximately 10% of reads will not be appropriately mapped simply due to gaps in the genome. Conversely, it also means that 90% will be correctly mapped.
The second potential pitfall, and which we have found more troublesome, lies in small scaffold sizes and the effect this has on identifying the genes associated with the called peaks. In a genome with many small scaffolds, a peak may be called that is orphaned from the gene to which it corresponds (Fig 1C). Additionally, if a break in the scaffold falls between a peak and the nearest gene, it is likely that the peak will be associated – not with its true nearest gene – but with its nearest gene existing on the same scaffold (Fig 1D). Although a large percentage of the X. laevis genome is contained on scaffolds larger than 50kb (currently 91.4%, and expected to increase to 94.7%), a more useful metric is the N50/L50 score, which describes the number (N) of scaffolds of length (L) that contain half the genome. For X. laevis v.6, the N50/L50 is 1102/700.7 KB, meaning that 1102 scaffolds of 700.7KB contain half the genome. The anticipated N50/L50 for v.7 is 648/1.1 MB, a considerable improvement meaning that fewer, larger scaffolds will contain half the genome, and making it less likely that peaks will be orphaned from their target genes. For comparison, the N50/L50 of X. tropicalis v4.1, found on the UCSC genome browser, was 272/1.56MB, which will continue to be better than the newer version of the X. laevis genome. Given that the genomes are divided into thousands of disconnected scaffolds, one major problem could be if many ChIP peaks appear on unannotated scaffolds. However, for both Stage 10 Smad2 ChIP and an H3K27ac ChIP (9) in X. tropicalis, we found that ~95% (4727 / 4952 for Smad2, 49080 / 51932 for H3K27ac) peaks were located within 1 MB of an annotated gene transcriptional start site from XGC, with similar results when using Refseq genes. Thus, our results have yielded much information even though the genomes are as of yet incompletely assembled.
6. Analysis
6.1 Sequencing platform considerations
A standard ChIP-SEQ analysis requires a sequenced ChIP library as well as a background library, which is usually input chromatin or IGG control ChIP. The most common sequencing platforms at the time of this writing are the Illumina GAIIx or HiSeq machines. Both platforms are capable of producing different read lengths, ranging from 36 to 100bp, as well as single or paired-end reads. For most ChIP applications, single-end 36 bp reads are sufficient. The primary advantage of longer and paired ends is increased mapability in unique regions, which may be beneficial if investigating the chromatin of repetitive genomic DNA. The major difference between these platforms is read throughput: the GAIIx routinely produces 30–50 million reads, while the HiSeq platform routinely generates 100–200 million reads per lane. For most ChIP experiments, 20–30 million reads is sufficient to produce a high quality library, although this depends on large part on the protein being investigated. For example, TF binding sites cover small punctuate sites (<300 bp) throughout the genome, and require less read coverage than histone modifications that cover broad (1 kb+) regions of the genome. While HiSEQ lanes are more costly, multiple samples can be “bar-coded” or indexed on the HiSEQ platform (consult Illumina for sequences of indexing primers). This way, multiple ChIP libraries can be sequenced on the same lane, making the effective cost far lower than the GAII without sacrificing read depth, since the HiSEQ platform yields much higher read count. We recommend anywhere from 4–6 samples be sequenced per HiSeq lane. Read indexing is usually performed by using Illumina adapter indexes or custom barcodes, but researchers should consult their sequencing facility.
6.2 Alignment
Because both RNA-SEQ and ChIP-SEQ libraries are double stranded DNA, they are handled by the sequencing platform in the same way. Sequencing centers usually generate a compressed “FASTQ” file, which includes all read sequences accompanied by quality score. Upon request they will usually align the reads to a user-specified genome. If not, there are multiple short-read aligners to accomplish this task. In Figure 2 we outline a general pipeline for sequence analysis, beginning with alignment. We use BWA (http://bio-bwa.sourceforge.net/), although Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) and SOAP (http://soap.genomics.org.cn/) are other popular aligners. For all of these programs, the original genome fasta file must be downloaded from the UCSC genome browser or Xenbase, and this file must be first be indexed by the aligner. Next, a single command is usually used to align the FASTQ file to the genome index, and the result is an alignment file in SAM format. The SAM format is widely adopted as the standard alignment format, and is often stored in compressed form as a BAM file. Importantly, the SAM output from an aligner may include redundant or unmapped reads, and these should be discarded. Programs such as Samtools (a basic toolkit for SAM manipulation)(http://samtools.sourceforge.net/) or Picard (which is more powerful but slightly more difficult to use)(http://picard.sourceforge.net/) should be used to “clean” SAM files prior to ChIP analysis, ensuring only uniquely mapping reads are utilized.
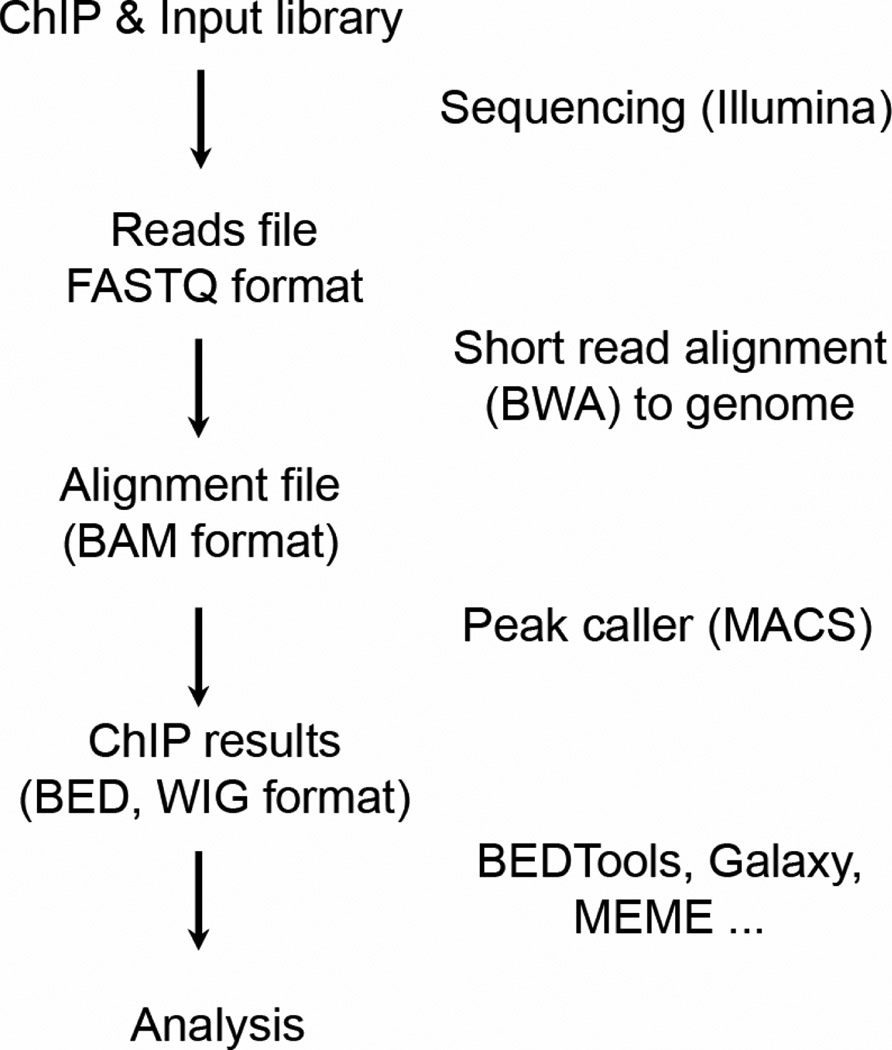
Figure 2.
Sequencing analysis workflow. An example workflow for sequencing data, alignment, peak calling, and analysis, including recommended programs for each stage.
6.3 Peak calling and peak inspection
Once a BAM file is generated for both ChIP and background samples, they can be used as input into a ChIP analysis program. Many programs exist for this purpose, and we recommend trying several as new and improved algorithms are constantly being developed. A major caveat when analyzing frog data is once again the scaffold arrangement. These gaps in the sequence break certain programs like QuEST (http://mendel.stanford.edu/SidowLab/downloads/quest/), but we found that the MACS program (http://liulab.dfci.harvard.edu/MACS/) works well with scaffolds when using the genome size set to 1.5e9. Most ChIP analysis programs will create a BED file that lists the coordinates of predicted ChIP peaks, as well as a WIG or BDG file that contains continuous ChIP coverage information over the genome.
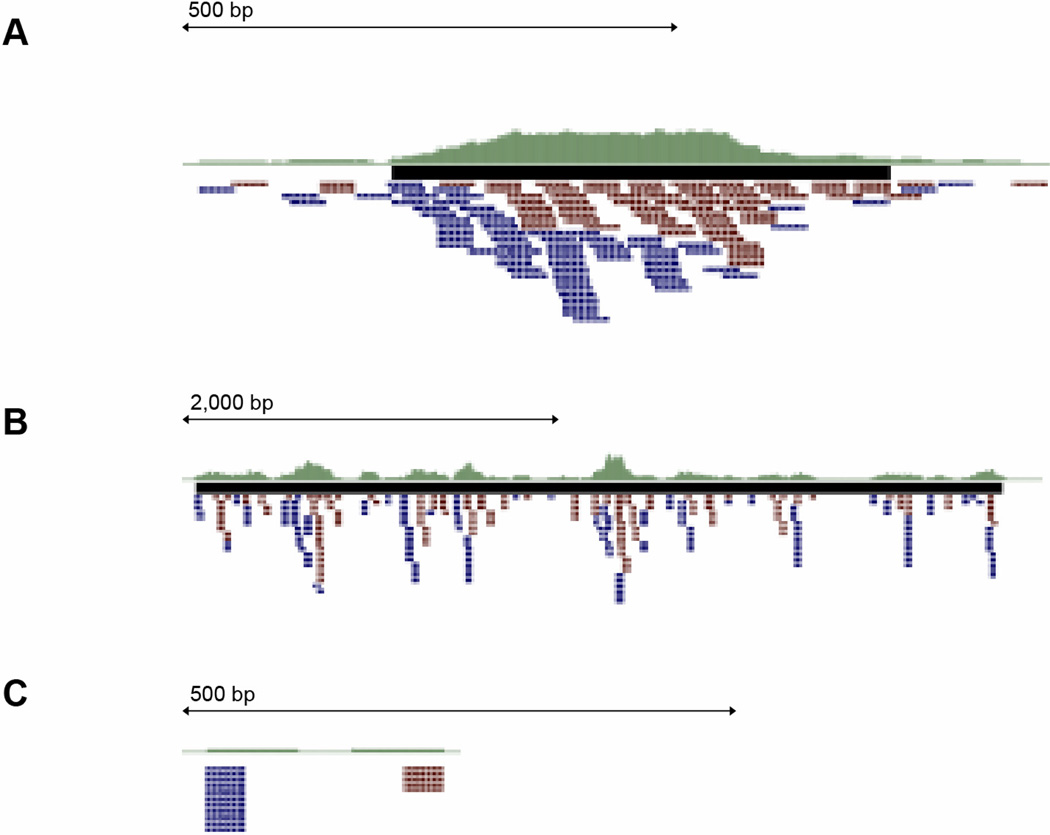
With all programs, the user should manually inspect predicted ChIP peaks with the raw reads, allowing for both evaluation of the quality of the data and for insight into threshold setting. The most reliable way to manual inspect the peaks is to upload the BAM file and any BED/WIG files as tracks in genome browser (e.g. using “add track” at the UCSC Genome Browser or Xenbase). At each predicted ChIP peak, the BAM track should display a smooth “pileup” of both positive and negative reads, with the width of the pileup dictated by the type of protein being detected. Typically transcription factor peaks will have a narrower peak (Figure 3A) and histone modifications a broader peak (Figure 3B). If there are <10 reads in most peaks, or if reads form a vertical pile rather than a smooth pileup (Figure 3C), or if reads are all coming from a single strand, then the settings should be adjusted to be more stringent (e.g. set a lower FDR or P value threshold). Conversely, if a region exhibits a characteristic pileup of reads but the program did not predict that region, the settings may be too stringent. If the majority of high-scoring peaks do not pass visual inspection, then the initial ChIP library was likely of poor quality. Another good metric of quality is to determine the % of uniquely mapped reads that fall within predicted peaks. This can be calculated by using BedTools (http://code.google.com/p/bedtools/) to count the number of reads from the BAM file that overlap the predicted peaks in the BED file (described below). A low fraction (< 10%) indicates that the library is noisy, and the ChIP may need to be repeated. Finally, researchers should also be aware of PCR “bottlenecking” where many reads pile in sparse vertical stacks rather than smooth piles across the genome (Fig 3C). This is indicative of either poor ChIP quality or low starting ChIP material when building the sequencing library, and results from PCR over-amplification. Overall, there are several specific features indicative of high quality ChIP libraries and we recommend that labs new to ChIP obtain a library of known quality for comparison.
Figure 3.
UCSC Screenshots of selected peaks. Green plot: The “Signal Per Million Reads” (SPMR) coverage density (WIG file) generated from MACS. Black bar: Predicted peak regions (BED file) from MACS. Red/blue tags: Aligned read data (BAM file). A) Characteristic transcription factor (Smad2) binding site, 300–500 bp wide, with a smooth pile of both + and − strand reads (blue and red, respectively) shifted by approximately the fragment length. B) Characteristic histone modification (H3K27ac) binding site, 2,000–4,000 bp wide with reads broadly distributed. C) Vertical “stacks” of multiple reads aligning to a single location, indicative of PCR over-amplification, usually from poor library quality or low starting input.
The number of peaks predicted for a library can range anywhere from a few hundred to tens of thousands. We rarely see libraries with over 100,000 predicted peaks. Some peak-caller programs such as MACS can also be used to determine whether adequate coverage has been obtained. For example, if a ChIP library was returned with 30 million reads, would resequencing the library to attain a total of 60 million reads identify more significant peaks? By randomly selecting a subset of reads, and re-running the ChIP program, one can determine whether adding more reads would be beneficial. MACS performs this automatically using the “--diag” mode, but essentially this mode predicts peaks using progressively larger subsets of the total reads (e.g. 20, 40, 60, 80, and 100%). If the number of peak predictions continue to increase substantially as the number of reads used increases, particularly from 80 to 100%, then more sequencing may be effective. However, if using 80% of reads identifies a similar number of peaks as 100%, then more reads are unnecessary. For some libraries, such as those with poor overall ChIP enrichment or if the protein of interest is widely bound in the genome (e.g. H3K9me3), attaining full coverage may be prohibitive. We usually find that peaks identified with further sequencing are usually those with low enrichment, so the most strongly enriched peaks will normally be found within 20–30 million reads.
6.4 Analysis tools suggestions
Once the user is assured of the quality of ChIP predictions, data analysis may proceed. The most standard analyses involve associating ChIP peaks with nearby genes, or determining the association of ChIP peaks across different libraries. In general, these analyses are performed using BED files. Annotated gene files, from Refseq or the Xenopus Gene Collection (XGC) can be downloaded in BED format from the UCSC Genome Table Browser. A number of programs exist to manipulate and compare BED files (determining overlap, adjacent elements, etc.). For most users we recommend using the online Galaxy suite of tools (https://main.g2.bx.psu.edu/), which offers a web interface for uploading and manipulating BED files. Downloadable programs such as BEDTools offer similar functionality with more customizability for those comfortable with command-line usage. Another common analysis is to predict sequence motifs enriched within a ChIP dataset. Many programs exist for this usage, but one of the most widely used is the MEME suite (http://meme.nbcr.net/meme/), which is capable of both de novo motif discovery (using Meme) and identifying instances of known motifs (using Fimo). Though a ChIP dataset may contain many thousands of peak predictions, we recommend using just the top 2000 highest-scoring peaks. These programs take as input the DNA sequence of all peaks in FASTA format, which can be generated using BEDTools or Galaxy.
Most analyses are readily performed on modern desktop computers. The most intensive step is often the alignment of reads to the genome, which can take several hours to a full day for moderately sized (20+ million reads) libraries. Most alignment tools, including BWA (using the –t option), can utilize multiple CPUs which are featured on most modern desktop computers. Peak calling may also take several hours, but most downstream analysis of BED files is relatively computationally unintensive. A major consideration is that sequence files and intermediate processed files are often several gigabytes each, and it is important to invest in sufficient hard drive storage space and backup solutions. If an adequately powerful machine is not readily available, remote servers such as Galaxy may provide a temporary solution, and many institutions now offer bioinformatics core servers that will usually have commonly used alignment software installed.
Example commands used for ChIP-Seq analysis
Using software: BWA 0.5.9, samtools 0.1.10, and MACS 1.4
To align reads to a genome “xenTro3.fa” downloaded from UCSC:
# creates a searchable index file for the genome
bwa index –a bwtsw xenTro3.fa
# aligns “reads.fastq” short-read file
# optional -t N where N is number of processors to use, ex) -t 4
bwa aln -q 10 xenTro3.fa reads.fastq >reads.sai
bwa samse xenTro3.fa reads.sai reads.fastq >output.bwa.sam
# Converts sam file to bam file, and only retains reads with quality scores of 1 or greater (scores of 0 indicate multiply mapped reads)
samtools view -S -b -q 1 output.bwa.sam >output.bwa.uniq.bam
# Calls peaks given uniquely aligned ChIP and control libraries (“chipReads.bam” and “inputReads.bam”).
macs14 -t chipReads.bam -c inputReads.bam -g 1.5e9 bam -n MyChIP
7. Conclusions
In this article we have supplied a general protocol for preparing ChIP samples in Xenopus using current methods, which are applicable to both Xenopus species and a wide variety of tissue types. We have offered our best current advice for navigating some of the specific challenges inherent to successful ChIP-SEQ in Xenopus, including sonication conditions, antibody choice and validation, and optimizing high-quality library preparation. We have also provided discussion of the current state of the Xenopus genomes and how we see this affecting ChIP in these species. While we hope that the approaches we provide here will continue to represent a useful starting point for analysis, we view the rapid changes in both UHTP sequencing technologies and Xenopus genomics resources with excitement and optimism, and expect that improvements will continue to arise over the next several years.
Acknowledgements
We are grateful to John Young of Richard Harland’s lab at UC Berkeley for advice on the strategy for controls using FLAG-tagged proteins. We also thank Taejoon Kwon of Edward Marcotte’s lab at the University of Texas, Austin and Adam Session of Daniel Rokhsar’s lab at UC Berkeley for information and discussion of the present state of the Xenopus laevis genome, and Duygu Ucar at Stanford for discussion of the impact of its stage of assembly on peak calling. We are indebted to the X. laevis Genome Consortium for releasing the genome prior to publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010 Apr 30;328:633. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkers RC, Jacobi UG, Veenstra GJ. Chromatin immunoprecipitation analysis of Xenopus embryos. Methods Mol Biol. 2012;917:279. doi: 10.1007/978-1-61779-992-1_17. [DOI] [PubMed] [Google Scholar]
- 3.Blythe SA, Reid CD, Kessler DS, Klein PS. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev Dyn. 2009 Jun;238:1422. doi: 10.1002/dvdy.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 2011 Aug 1;25:1654. doi: 10.1101/gad.16800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982 Oct;30:687. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 6.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982 Oct;30:675. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 7.Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS. An essential role for transcription before the MBT in Xenopus laevis. Dev Biol. 2011 Sep 15;357:478. doi: 10.1016/j.ydbio.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell JSaD. Molecular Cloning. A laboratory Manual. 3rd edition. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 9.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012 Apr 12;484:251. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker JC. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]