Figure 2. Statistical analysis and validation: Histogram for the number of significant genes and their fold changes from the actual combination of Bead Chips compared to 20 random combinations (p<0.05) are shown for both (A) sex and (B) Genotype.
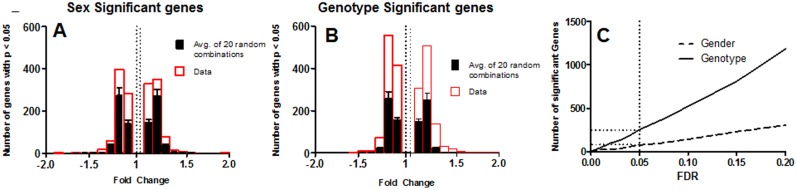
This shows the number of significantly identified genes in the actual combination is higher than number of false positives expected, even at low fold change. Number of significant genes as a function of FDR value is shown, at every FDR level genotypic variation is larger than the (C) sex variation. When corrected for multiple comparison (FDR) no significant fold changes are identified in any of the random combination until FDR = 0.20. Significant genes were calculated using Partek Genomics Suite, using three way ANOVA model on filtered dataset for signal intensity and batch effect (13, 155 probe sets of 25179) and p value <0.05, corrected for FDR <0.05.
