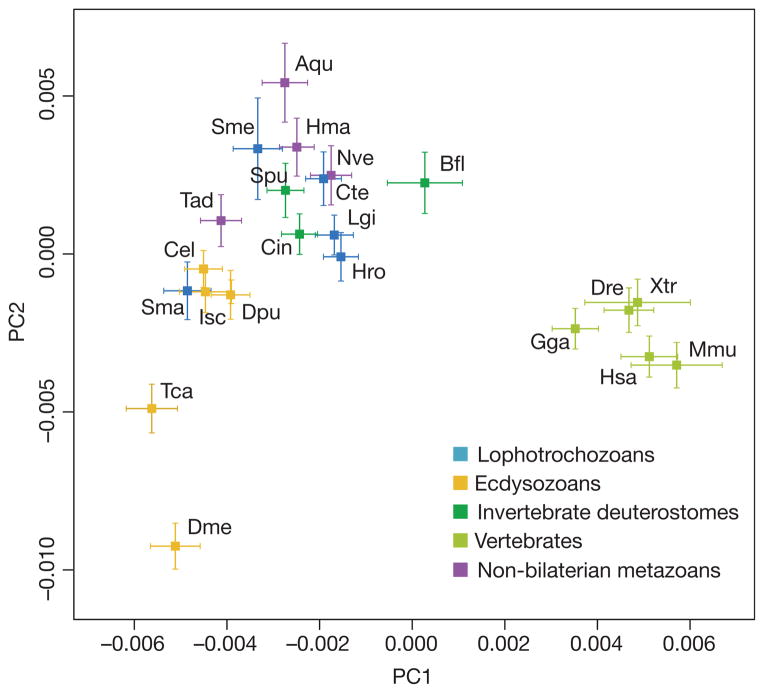
Figure 2. Clustering of metazoan genomes in a multidimensional space of molecular functions.
The first two principal components are displayed, accounting for 20% and 15% of variation, respectively. At least three clusters are evident, including a vertebrate cluster (far right), a non-bilaterian metazoan, invertebrate deuterostome or spiralian cluster (centre, top), and an ecdysozoan group (lower left). Drosophila and Tribolium (lower left) are outliers. Aqu, Amphimedon queenslandica (demosponge); Bfl, Branchiostoma floridae (amphioxus); Cel, Caenorhabditis elegans; Cte, Capitella teleta (polychaete); Cin, Ciona intestinalis (sea squirt); Dme, Drosophila melanogaster; Dpu, Daphnia pulex (water flea); Dre, Danio rerio (zebrafish); Isc, Ixodes scapularis (tick); Gga, Gallus gallus (chicken); Hsa, Homo sapiens (human); Hma, Hydra magnipapillata; Hro, Helobdella robusta (leech); Lgi, Lottia gigantea (limpet); Mmu, Mus musculus (mouse); Nve, Nematostella vectensis (sea anemone); Sma, Schistosoma mansoni; Sme, Schmidtea mediterranea (planarian); Spu, Strongylocentrotus purpuratus (sea urchin); Tad, Trichoplax adhaerens (placozoan); Tca, Tribolium castaneum (flour beetle); Xtr, Xenopus tropicalis (clawed frog).