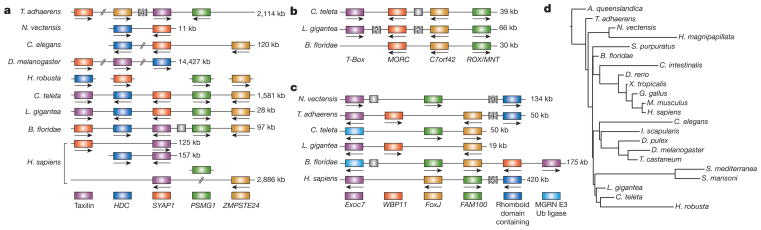
Figure 5. Examples of conserved orthologous gene clusters.
a–c, Clusters of linked genes across diverse species. Within each panel, genes in the same colour are members of the same orthologous group, with the gene identifiers of defining members of the group indicated: C7orf42, (human) chromosome 7 open reading frame 42; Exoc7, exocyst complex component 7; FAM100, family with sequence similarity 100; FoxJ, forkhead box protein J; HDC, histidine decarboxylase; MGRN E3 Ub ligase, mahogunin ring finger E3 ubiquitin ligase; MORC, MORC family CW-type zinc finger; PSMG1, proteasome (prosome, macropain) assembly chaperone 1; ROX/MINT, Max-binding protein family member; SYAP1, synapse-associated protein 1; WBP11, WW domain binding protein 11; ZMPSTE24, zinc metallopeptidase, STE24 homologue. Scaffold positions for all displayed linkages are listed in Supplementary Note 7.2. d, Cumulative rates of microsynteny change plotted on a fixed metazoan tree topology. Branch lengths are proportional to the number of inferred genomic rearrangements. A. queenslandica, Amphimedon queenslandica; C. intestinalis, Ciona intestinalis; D. melanogaster, Drosophila melanogaster; D. pulex, Daphnia pulex; D. rerio, Danio rerio; G. gallus, Gallus gallus; H. magnipapillata, Hydra magnipapillata; I. scapularis, Ixodes scapularis; M. musculus, Mus musculus; N. vectensis, Nematostella vectensis; Ub, ubiquitin; X. tropicalis, Xenopus tropicalis.